Deep-learning-based prediction of living cells mitosis via quantitative phase microscopy  Download: 557次
Download: 557次
1. Introduction
Cell morphology provides meaningful information on cell physiology and has a wide range of applications in the monitoring of cell status during biological processes. The most important cell events, such as cell division, significantly change the properties of a whole cell. For example, during cell division, an ancestor cell shrinks and becomes circular and then splits into two daughter cells. Accurate biophysical parameter measurements during mitosis of adherent cells are particularly necessary, as cells undergo, both in culture and in tissues, important changes in morphology[1]. However, mitosis events may often appear randomly in biological systems and vary depending upon the cells growth conditions. Long-term monitoring of the cellular morphological dynamic process is usually demanding for obtaining cell mitosis videos.
There have been a number of methods for cell mitosis identification using deep learning combined with optical microscopy over the past few years[2
As a label-free imaging technique, quantitative phase microscopy (QPM) enables rapid acquisition of overall cellular information on both the morphology and refractive index for living single cells[810" target="_self" style="display: inline;">–
In this Letter, we present an approach based on QPM and DCNN to predict mitosis and non-mitosis at the single-cell level. Here, a through-focus QPM system based on the transport of intensity equation (TIE) described previously is used for obtaining the phase images of living cells[19]. Potential mitotic cells are cropped from the overall phase images and divided by the experienced biologist into mitosis and non-mitosis based on morphology variation. Then, the labeled quantitative phase images of individual mitotic candidates are collected as a dataset for DCNN training. For comparison, labeled intensity images of individual mitosis and non-mitosis are also collected for training DCNN. The DCNN trained by phase images achieves a higher average accuracy and F1 score than the DCNN trained by intensity images on automated classification of mitosis and non-mitosis.
2. Materials and Methods
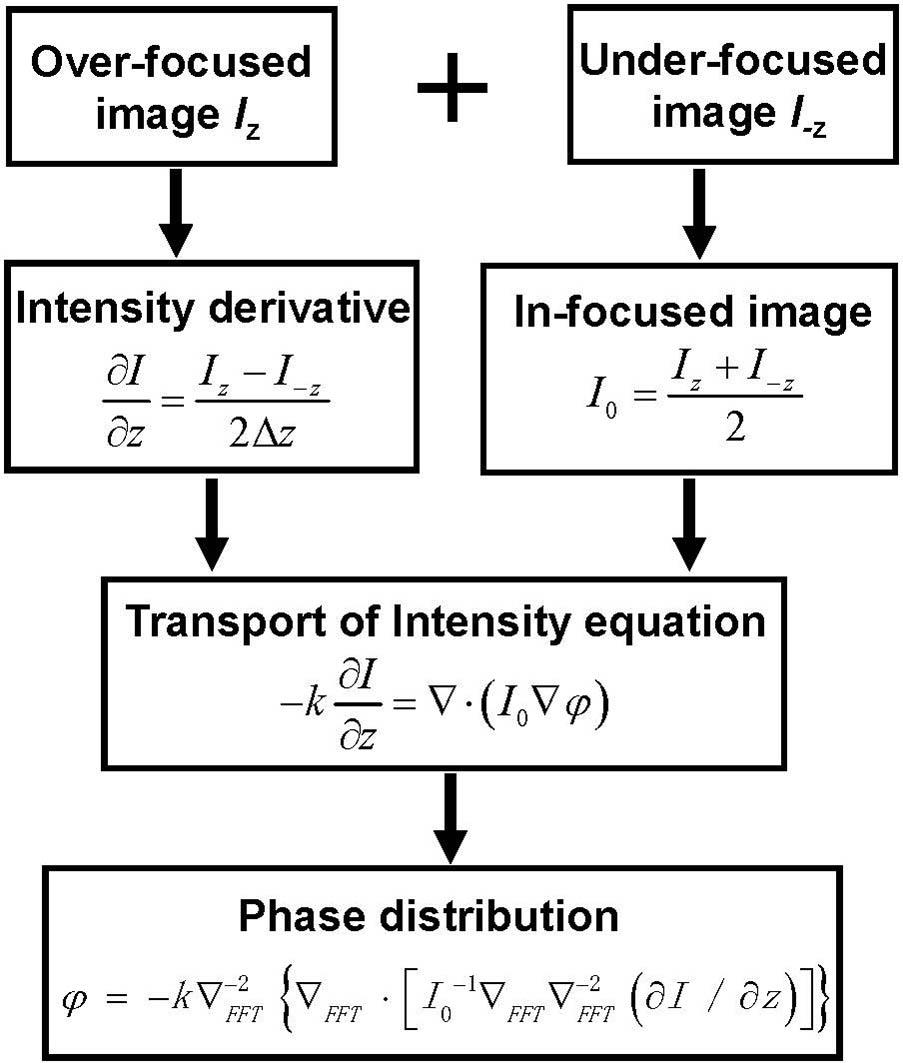
Figure 1 shows the flowchart of phase recovering based on the through-focus QPM. Here, two out-of-focus images are used, which are over- and under-focused with the same focal distance away from the in-focus plane, respectively. According to the finite difference, the derivative of the image intensity with respect to the optical axis can be approximated with the over- and under-focused images. The in-focus intensity image is computed as the average of the two symmetrically defocused images[20,21]. Then, based on the calculated in-focus intensity and intensity derivative, quantitative phase image can be uniquely determined by solving the TIE under proper boundary conditions based on fast Fourier transform[22,23].
The QPM system is shown in Fig. 2(a). This experimental setup is built on an inverted bright-field microscope upgraded with a flipping imaging module[19]. A single image at the input of this flipping imaging module can be doubled as two laterally distributed images on the CCD camera (Imaging Source SVS16000MFGE, , pixel size 7.44 µm, 15 frames/s). Next, using a 5 µm pinhole as the object, the two laterally separated images can be fine-tuned as in-focused by translating the M2 axially to a certain plane. Then, by translating M2 Δz away from the plane, a pair of in-focus images and defocused images can be created simultaneously. By properly translating the microscope stage along the optical axis, ensuring that the sample is precisely located at the central position between over- and under-focused planes, two images with defocus distances of can be captured.

Fig. 2. (a) Experimental setup. D, diaphragm; BS, beam splitter; RR, retroreflector; M2, mirror 2. (b) 2D images of source pattern and corresponding phase transfer function for different coherence parameters. (c) Original lateral-flipped images in single shot. (d) Recovered phase map of the cell marked in the red box in (c). (e) Phase stability measurement during the 150 min experiment in the white box marked in (d).
The QPM system employs Kohler illumination, whose spatial coherence is represented by coherent parameter S, the ratio of the numerical aperture of the condenser to that of the objective. The simulated phase transfer function for various coherent parameters is shown in Fig. 2(b), and the value for S is maximized by ∼0.6[2426" target="_self" style="display: inline;">–
This system is calibrated using a plano-convex microlens array (OPTON, MLA-2R250, 250 µm pitch) by comparing the measured height with the manufactory value to confirm the validity of this approach. In the measurement of living osteoblastic cell specimens, low cell density covering only 30% to 40% of the chamber is adopted to avoid cell overlapping and ensure homogenous background. A objective is used for living cell imaging.
Figure 2(c) shows the original lateral-flipped images of living osteoblastic cells, and Fig. 2(d) gives the phase of the cell marked in the red box. An area of pixels in the top-left corner of the cell [indicated by a white square in Fig. 2(d)] is selected, and the average phase fluctuations in this area are calculated over 150 min. The standard deviation of the graph shown in Fig. 2(e) is only 0.0128 rad, corresponding to 0.002λ.
Mitosis events may often appear randomly in nature and vary depending upon the conditions under which the mitosis events are usually sparsely distributed. Considering that the cell lifecycle is around 20 h, after being taken outside the clinostat, the living cells are placed on the microscope stage under monitoring for a whole day[14]. Some cells in the observation window undergo the representative lifecycle morphology changing process. For these cells, the time-lapse phase images during the division stage are cropped from the original whole phase images.
As shown in Figs. 3(a)–3(d), the phase images clearly reveal the cell morphological changes during different phases of mitosis over the course of 30 min. At prophase and metaphase, the mitotic cells retract and round up. At anaphase, the sister chromatids are separated and pulled toward the spindle pole, and the poles of the cell scale out at the same rate simultaneously. At telophase, the ancestor cell splits into two daughter cells, which look like the shape ‘8’. In this stage, mitosis is easy to identify by observing the visual appearance. Compared with the four distinct stages experienced by mitotic cells, non-mitotic cells only continue to retract and round up in morphology at a slow speed over several hours, as shown in Figs. 3(a) and 3(b).

Fig. 3. (a), (b) Representative phase and corresponding intensity images of mitosis. (c), (d) Representative phase and corresponding intensity images of non-mitosis.
Obviously, it is difficult for researchers to figure out whether rounding-up cells is in the mitotic events or non-mitotic process using a single phase image. DCNN, which is capable of discovering effective representations from image data in a self-taught manner, is further proposed for subsequent automatic identification of cell states[27]. The time-lapse phase images of mitotic cells during prophase, metaphase, and anaphase, together with the dynamic phase images of non-mitotic cells, are used for the training of DCNN afterwards.
DCNN is built by modifications of AlexNet, which has been pretrained using millions of high-resolution images in ImageNet and is able to be fine-tuned to fit our dataset using transfer learning[27]. The fine-tuned modifications using transfer learning are as follows[28].
The detailed architecture of DCNN after transfer learning is illustrated in Fig. 4. The phase image of a single cell is processed by five convolution layers each convolution layer followed by a rectified linear unit (ReLU), three 2 × 2 max pooling layers with stride 2, and two fully connected layers each followed by an ReLU. At the end of the overall network, a softmax function is used for binary classification.

Fig. 4. Workflow diagram and detailed architecture of DCNN for classification of mitosis and non-mitosis.
The collected potential mitosis dataset contains 40 mitotic cells and 45 non-mitotic cells. For both mitotic and non-mitotic cells, 80% of this dataset is used for training, and 20% is used for validation. Data augmentation techniques are utilized to reduce overfitting and increase accuracy. The data augmentation generates virtual images using flipping and rotation in four different angles (0°, 90°, 180°, 270°) to help increase the robustness of DCNN.
To remove the effect of phase information on the performance of DCNN, the dataset consisting of intensity images is used to train DCNN for comparison. As a result, DCNNs are separately trained by phase images and intensity images, respectively. Training loss, validation loss, training accuracy, and validation accuracy are calculated every 0.2 epoch during the training process of 50 epochs. The batch size and initial learning rate are set as 5 and 0.0005, respectively. The data is processed using MATLAB based on CPU computing, and the training process of 50 epochs takes 19 h.
3. Results and Discussions
The performance (loss and accuracy) of DCNN trained by phase images throughout the entire training and validation process is illustrated in Figs. 5(a) and 5(b). The loss and accuracy in validation start to remain steady after 3 epochs. After 50 epochs, the network achieves its final accuracy of 100%. Compared with the performance of DCNN using phase images as input, DCNN trained by intensity images demonstrates a detrimental effect on the performance shown in Figs. 5(c) and 5(d). The reduced validation accuracy and the increased validation loss indicate that the network trained by intensity images shows an impaired performance.
Figures 6(a) and 6(b) show the prediction ratio of mitosis and non-mitosis based on DCNN trained by phase and intensity images, respectively. As can be seen from Fig. 6(a), DCNN trained by phase images performs with almost excellent accuracy (0% false negative error) on the discrimination between mitosis and non-mitosis. The prediction results in Fig. 6(b) indicate that an error occurs when DCNN is trained by intensity images. Some cells from the non-mitosis class are wrongly classified as mitosis.

Fig. 6. Performance and t-SNE visualization of the DCNN trained by phase and intensity images, respectively. In t-SNE visualization, number 1 represents mitosis and number 2 represents non-mitosis.
To further interpret and visualize the classification performance of the two trained DCNNs on single-cell data, a nonlinear dimension reduction technique, -distributed stochastic neighbor embedding (-SNE) is applied to the activation of individual neurons in the last fully connected layer. Using -SNE, the input images at the head of the network are transformed into representative spots in a two-dimensional coordinate plane, where cells with similar features are close to each other, and cells from different classes spread far away from each other. The -SNE visualizations of the two DCNNs are shown in Figs. 6(c) and 6(d). The distinct separation between mitosis and non-mitosis in Fig. 6(c) indicates optimal representation of DCNN trained by quantitative phase images.
Figure 7 shows the overall classification performance (accuracy, precision, recall, and F1 score) of the two DCNNs by applying five-fold cross validation on the dataset. The average accuracy, precision, recall, and F1 score of the DCNN trained by phase images are 98.9%, 100%, 94.8%, and 97.4%, respectively. The DCNN trained by intensity images leads to poorer performance, when the accuracy, precision, recall, and F1 score of the classification are 89.6%, 72.6%, 100%, and 83.8%, respectively. Earlier publications also reported that machine learning classifiers trained by phase images outperform the classifiers trained by intensity images[17,18,29]. Compared with quantitative phase images reflecting the intracellular refractive index, the intensity images only offer rough information on cell contour.

Fig. 7. Overall classification performance for DCNN trained by phase images, DCNN with intensity images and random forest trained by dry mass, respectively. (a) Accuracy. (b) Precision. (c) Recall. (d) F1 score.
From the above results, we reason that the quantitative phase signal, as an important functional characteristic of cell status, is capable of discerning the cell mitotic state in combination with DCNN. Due to the morphological similarities between mitosis and non-mitosis, the information involved in intensity images is insufficient for cell status discrimination.
In particular, an important cell biophysical parameter termed cell dry mass, was previously utilized for various purposes, including phenotyping dynamics during mitosis and cell cycle[1,13,14]. The dry mass of a living cell is calculated from surface integral of the phase information, as detailed in earlier reports[9]. The computed dry mass dataset of mitotic and non-mitotic cells is used to train a random forest classifier. Using five-fold cross validation, the overall classification results are shown in Fig. 7. In this case, the accuracy, precision, recall, and F1 score are 89.3%, 87.7%, 85.7%, and 86.7%, respectively. The performance of random forest based on dry mass is comparable to the performance of DCNN trained by intensity images. While in contrast to the DCNN trained by phase images, the random forest shows a relatively poorer performance, which means that the quantitative phase distribution plays an important part in the classification of mitosis and non-mitosis.
Table 1 further compares the performance of three different network settings. “AlexNet” is a CNN used for classification of mitotic cells in histopathological breast cancer images[6]. “CNN only” is the CNN using phase contrast images as input for identification of mitosis[2]. This comparison validates that the DCNN trained by phase images produces more superior results than the network using histopathological breast cancer images or phase contrast images for mitosis detection.
Table 1. Comparison of Accuracy and F1 Score for Different Classifiers
|
4. Conclusion
We have demonstrated the performances of deep learning based on quantitative phase images and intensity images when dealing with the classification of mitosis and non-mitosis. The DCNN generated from quantitative phase images shows better performance in binary classification of mitosis and non-mitosis in comparison with the DCNN generated from intensity images. Based on these results, it can be inferred that the quantitative phase images provide a means to identify the subtle differences and slight variations between mitosis and non-mitosis at the single-cell level. In further combination with deep learning, QPM can be developed into a useful tool to improve the performance on automatic analysis of living cell status noninvasively, such as cell death, cell cycles, and cells under different experimental conditions.
[5] Ş. Öztürk, B. Akdemir. A convolutional neural network model for semantic segmentation of mitotic events in microscopy images. Neural Comput. Appl., 2018, 31: 3719.
[23] M. Beleggia, M. A. Schofield, V. V. Volkov, Y. Zhu. On the transport of intensity technique for phase retrieval. Ultmi, 2004, 102: 37.
Ying Li, Jianglei Di, Li Ren, Jianlin Zhao. Deep-learning-based prediction of living cells mitosis via quantitative phase microscopy[J]. Chinese Optics Letters, 2021, 19(5): 051701.