Resolution-enhanced intensity diffraction tomography in high numerical aperture label-free microscopy  Download: 689次
Download: 689次
1. INTRODUCTION
The refractive index (RI) of biological cells and tissues plays a crucial role in biomedical imaging as it captures the physiological characteristics and morphological features of a sample [1–3]. Imaging 3D biological samples with high resolution, however, remains a challenging task. Fluorescence-based 3D imaging methods, such as confocal microscopy [4] and multiphoton microscopy [5], already provide excellent optical sectioning and enable super-resolution 3D volume imaging of nanoscale subcellular structures. Unfortunately, the exogenous labels (e.g., dyes and fluorophores) in these techniques can be phototoxic and cause photobleaching due to the absorption of the excitation light [6] and can artificially alter the sample’s behavior and cellular structure [7]. Here, we introduce a resolution-enhanced intensity diffraction tomographic (reIDT) technique based on a conventional bright-field microscope and a high-density LED array illumination unit with sparse annular illumination. The imaging method is label-free, motion-free, fast, and enables the high-resolution quantitative recovery of 3D intrinsic structural features at the single cell level.
Quantitative phase imaging (QPI) [3] is an often-used technique to recover biological cells with high contrast in their natural states. QPI reconstructs a quantitative image of the sample-induced two-dimensional (2D) optical path thicknesses or phase delay based on interferometry [8–11] and noninterferometry [12–18] methods. However, these phase maps evaluate the sample’s total optical delay along the integrated direction over a thin layer and provide limited volumetric information about the object’s internal structure. To capture this information and evaluate thicker specimens, tomographic methods recovering 3D information are required. Recently developed 3D RI tomography techniques enable the reconstruction of the sample’s 3D RI distribution to visualize intracellular structures, and the related 3D imaging techniques can be largely categorized into two classes, interferometry-based and intensity-only methods.
Optical diffraction tomography (ODT) is the most widely used interferometry-based 3D QPI technique [19,20]. Standard implementations of ODT use either a rotating sample [21] or a scanning laser beam [22–24] to capture the angle-specific scattering arising from the sample. A 2D complex scattered field containing the object’s phase and amplitude information is directly recorded as digital interferograms under various illumination angles, and a tomographic reconstruction algorithm is applied to recover the sample’s 3D RI distribution. The interferograms are captured using either an off-axis [20–24] or common path [25–27] holographic microscope, and numerous ODT approaches [22–24] have been applied in biomedical imaging for achieving quantitative, high-resolution 3D imaging. Hence, ODT can avoid the complex sample-preparation protocol and photobleaching effects of fluorescence while still recovering a sample’s 3D morphology, and this label-free 3D reconstruction method has found great success in visualizing individual unlabeled cells with quantitative refractive indices.
For intensity-only techniques, existing IDT techniques involve taking multiple defocused intensity images under symmetric [28–31] or asymmetric [32,33] illuminations, and the 3D RI map is recovered using deconvolution [34]. Techniques such as differential interference contrast acquire through-focus intensity stacks to extract the 2D phase information at multiple focus planes, and the 3D RI distribution is reconstructed using gradient phase information [35]. Recent works further incorporate modulated partially coherent sources (e.g., annular shape and Gaussian shape) to enhance the response of the 3D transfer function for noise-robust 3D tomographic results [30,31]. However, these methods often require sample or objective mechanical scanning along the axial direction to capture a through-focus intensity stack. This scanning limits the setup’s acquisition speed and requires large datasets with hundreds of intensity frames for object recovery. Alternatively, IDT approaches have been developed allowing 3D RI information recovery using angled illumination-only intensity measurements without any mechanical scanning [36–42]. These methods apply single-scattering approximation-based models enabling direct 3D object reconstruction with a closed-form solution [39–41]. Iterative algorithms based on multiple scattering models using the multislice beam propagation method have also been developed to recover strongly scattering samples [37,43,44]. One drawback of most of these existing techniques is the large number of required intensity measurements that consist of hundreds of intensity images per volume. The iterative reconstruction algorithm further prolongs the whole 3D imaging process and can become computationally expensive.
Here, we present a reIDT setup that utilizes a high-density LED array hardware and a high numerical aperture condenser to significantly improve the achievable resolution of the single-scattering-based IDT technique using a larger objective (). Compared to existing single-scattering-based IDT techniques [39–41], we improve the imaging resolution to near-diffraction-limited lateral resolution of 346 nm. Specifically, the achievable axial resolution is extended from 3.4 μm to 1.2 μm enabling single-cell-level 3D structure imaging. By introducing a condenser lens to the setup, reIDT improves the illumination efficiency and reduces the system’s required exposure time. The annular illumination pattern optimally fitted with the circular microscope pupil also reduces the image requirement to improve the system’s speed. Finally, we show that the present technique provides high-resolution, volumetric, subcellular information and morphology analysis on multiple biological samples. We recover macroscale cellular membrane structures, subcellular organelles, and micro-organism tissues to demonstrate that our technique has potential as a fast and high-resolution 3D imaging tool for biological samples.
2. THEORY AND METHOD
2.1 A. Principle of reIDT
As depicted in Fig. 1(a), the proposed reIDT imaging system contains a high-density LED array illumination unit and an Abbe condenser attached to a commercial bright-field microscope. By placing the LED array in the front focal plane of the condenser lens, each LED source located at the condenser aperture diaphragm provides spatially partially coherent illumination in the form of a plane wave at a unique illumination angle on the sample. The annular pattern is displayed on the LED array, and ring-shape illumination (radius , illumination numerical aperture ) is matched with the objective lens’s pupil aperture properly (i.e., ), as illustrated in Fig. 1(b). The specimen is placed in the objective’s focal plane and illuminated by each LED in the ring array. The intensity from each illumination is recorded on a camera which captures the interference of the object scattered field and the unperturbed illuminating field under the single-scattering approximation [Fig. 1(c)].
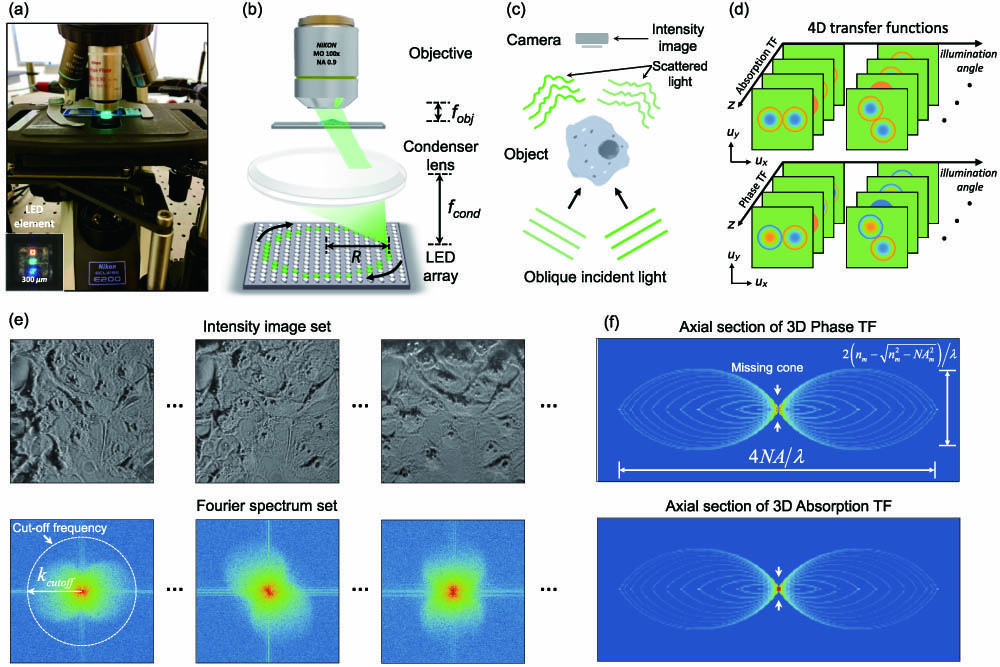
Fig. 1. System illustration of reIDT technique. (a) Photograph of reIDT setup. The setup consists of a standard microscope equipped with a high-density LED array and a high numerical aperture condenser. (b) LED array illumination unit is placed in the front focal plane of the condenser lens. The radius
By quantifying the Fourier space information under the first Born approximation, the phase and absorption transfer functions (TFs) of the IDT linear model can be derived [39], and the object’s complex permittivity distribution can be linearly related to the measured intensity spectrum. The 4D TFs in Fig. 1(d) contain slice-wise (2D) phase and absorption transfer functions at different depths for each illumination angle, and the exact form of the TFs can be found in Eq. (5) and Eq. (6). Importantly, since the TFs are confined to each slice, the scattering information from different sample slices is decoupled. This decoupling allows us to use a computationally efficient, slice-based deconvolution procedure to reconstruct the cross-sectional RI distribution one slice at a time. The raw intensity image set is captured by varying the illumination angles, and each intensity spectrum of the obliquely illuminated images exhibits a pair of circular Fourier regions. The pupil function filters the incoming scattered field to this circular shape, while the oblique illumination’s transverse frequency and its conjugate translate the scattered field in opposite directions in the frequency domain [Fig. 1(e)]. The lateral cutoff frequency envelope is indicated by the white dashed circle with the LED source’s illumination angle () in Fig. 1(e). By combining the LED position self-calibration algorithm, the accurate transverse frequency of incident illumination can be obtained and used for modeling the 4D TFs. Finally, a fast and efficient slice-wise 3D deconvolution algorithm with a closed-form solution [Eqs. (7) and (8)] is implemented to recover the sample’s complex 3D RI distribution. Since the IDT model is only valid for bright-field measurements, the system still suffers from the missing cone problem in the axial direction, resulting in the axial elongation present in the reconstruction. Figure 1(f) illustrates the frequency support region of the proposed reIDT method in k-space.
In practice, using a high-NA objective and illumination scheme cannot assume paraxial imaging conditions. As proposed by Sheppard and Gu et al. [45–47], high-angle microscope systems usually incorporate an apodization function introduced by the system satisfying the sine condition. Thus, the investigation of the objective aperture function modified with an apodization function instead of ideal step function should be our future work.
2.2 B. LED Position Self-Calibration
When reconstructing the object, the illumination angles of the ring geometry setup must be known exactly to generate a 4D TF stack matching the acquired dataset. Any small alignment variations in the experimental setup deviating from the expected illumination angles will misalign the Fourier spectrum information and induce poor-quality reconstructions. Especially for high-NA imaging situations [48], inaccurate estimations of illumination angles will degrade the final imaging resolution and axial reconstruction depth range under higher-NA objective with smaller depth of focus (DOF). To prevent this, we first perform a numerical self-calibration procedure to obtain the incident illumination’s transverse spatial frequency. We first adjust the radius of annular pattern on the LED array to approximately match the objective pupil, and 48 intensity images are captured under each illumination angle in the annular pattern. A self-calibration algorithm is then required to correctly obtain the illumination angles from the initial estimate.
This self-calibration process utilizes a previously developed algorithm [49] with geometric constraints for accurate object recovery. The LEDs are assumed to be equally spaced on a rectangular matrix and obey a pre-defined annular geometry. With these constraints, the positions of LEDs on the annular pattern can be expressed as

Fig. 2. Demonstration of the proposed self-calibration method on an LED matrix with annular illumination pattern. (a), (b) The LED positions from manual alignment (termed uncalibrated, marked in blue star), initial guess of calibration (termed Init-calibrated, marked in red dot), and calibrated spatial frequency positions of LEDs (green triangle), plotted in the spatial frequency coordinates. (c), (d) 1D abscissa and the ordinate spatial illumination frequency plots of each LED on the annular pattern. Uncalibrated spatial frequency positions
From these angle estimates, our algorithm obtains a better initial guess () from the algorithm in Ref. [49] to provide initial LED position estimates shown as the red dots in Fig. 2(a). In practice, the algorithm in Ref. [49] is subject to noise and angle mismatches that can place LEDs outside of the square grid of the LED matrix. We define the following global factors to determine the positions of the LED pattern elements: rotation factor ; shift factors and along , ; effective radius of objective aperture ; and pincushion distortion factors and . By substituting Eq. (2), the final calibrated LED positions and are parameterized as
2.3 C. Reconstruction Algorithm
After calibrating the illumination, the reconstruction algorithm combines all the intensity images to estimate the sample’s 3D complex permittivity contrast (i.e., phase and absorption). In the IDT forward model, a 3D sample is discretized into a stack of 2D sample slices. Following the IDT derivation [39–41], the 3D phase and absorption TFs can be derived slice-by-slice as a function of the illumination angle. The absorption and phase transfer functions (ATF and PTF, respectively) both contain a pair of circular Fourier regions describing the scattered field’s information and its complex conjugate [Fig. 1(d)]. The analytical expression of the phase TF () and absorption TF () can be expressed as
To achieve 3D RI reconstruction, reIDT solves an inverse problem through deconvolution. First, each intensity image is processed to remove the background. Next, the Tikhonov regularized deconvolution is performed to reconstruct the object’s real and imaginary RI. The main idea of this slice-wise deconvolution process is to replace the continuous integration along the axial direction by a discrete sum over the slice index [39]. The slice spacing is chosen to match the DOF of the imaging system during the computation. The achieved axial resolution is characterized by analyzing the reconstructed stack and is found to approximately match with the diffraction limit, set by corresponding full period resolution at the Nyquist–Shannon sampling limit, where is the RI of the surrounding medium. For the high-NA situation in this work, the reconstruction depth range limitation is smaller than the DOF of a lower-NA objective due to the smaller DOF of the higher-objective NA and more sensitive to high-angle illumination misalignment. These closed-form solutions for phase and absorption at each axial slice are
3. EXPERIMENTAL SETUP
Our reIDT system consists of an upright bright-field microscope (E200, Nikon) with a commercial high-density surface-mounted LED array in the front focal plane of the condenser as the illumination source shown in Figs. 1(a) and 1(b). The green source’s semiconductor die is located in the LED’s center position with a size of 150 μm. Thus, the LED source can be approximated as a spatially coherent source, and the light emitted from the condenser lens for a single LED can be nearly treated as a plane wave. Each LED can be controlled individually to illuminate the sample from a unique angle and approximately provides spatially coherent quasi-monochromatic illumination with central wavelength , bandwidth. The distance between LED elements is 1.25 mm, and the original resolution of this commercial LED array is . Due to the limited diameter of the circular condenser aperture, only a fraction of the whole array () is used in the experiment. The LED array’s center is aligned with the microscope’s optical axis and the annular illumination ring is matched with the perimeter of the objective lens’ numerical aperture.
All experiments in this work were conducted using a microscope objective (0.9 NA, CFI Plan Fluor, Nikon). Images were taken with an sCMOS camera (PCO.Panda 4.2, 6.5 μm pixel size), which is synchronized with the LED source to provide camera-limited acquisition speed. Moreover, the LED array is driven dynamically using an LED controller board built in-house using a field-programmable gate array (FPGA, EP4CE10E144) unit. For the image acquisition and data processing time, we capture 48 intensity images with pixels (2 pixels binning for 6.5 μm pixel size) and a 20 ms exposure time. This exposure time can be shortened and the total LED number can be downsampled to further improve acquisition speeds depending on the experiment. All the experimental data is processed in MATLAB (MATLAB 2018b) with a personal computer (Intel Core , 3.6 GHz, 128 GB DDR4 RAM). The required time to reconstruct a RI voxel stack is approximately 20 s.
4. RESULTS
4.1 A. 3D Absorption Reconstruction of USAF Target
We first demonstrate the imaging performance of our reIDT platform experimentally by measuring a 1951 USAF resolution target (Ready Optics Company, USA). The 3D absorption reconstruction of the resolution target is shown in Fig. 3. Even though the target was placed horizontally on the sample stage of the microscope, the lateral and axial imaging ability can be characterized using resolution elements and the axial full width at half-maximum (FWHM). Figure 3(a) shows the raw full field-of-view (FOV) image of the absorption USAF object under oblique illumination from the annular LED with the central features (white box) extracted and shown in Figs. 3(b) and 3(c). For the raw intensity images, the theoretically achievable resolution is limited by the incoherent diffraction limit (half-period width 151 nm, effective , and 0.9) which is larger than the camera pixel size (130 nm at the object plane), and thus we do not need to consider pixel aliasing effects. This intensity dataset was used to implement 3D tomography based on reIDT, and Figs. 3(b) and 3(c) present the smallest elements (Groups 10 and 11) in the central slice of the 3D absorption stack of the USAF target. The subregion of resolution gratings in Group 11, horizontal Element 4 is enlarged revealing that the highest resolution achieved is 346 nm of full periodic grating pattern distance according to the standard resolution metric reported in Ref. [50] (bar width 173 nm, Group 11, horizontal Element 4).

Fig. 3. 3D absorption reconstruction of the USAF target. (a) Raw full-FOV image of the absorption USAF object under oblique illumination from the annular LED. (b), (c) Enlarged central groups of resolution features in the central slice of the 3D absorption stack of the USAF target. (e) Multiple axial sections of USAF 3D absorption reconstruction at different planes and cross-section intensity profile plot of the recovered USAF target. (f) 3D volume-rendered view of the reconstructed absorption distribution. Additional cross-sectional reconstruction and 3D volume rendering from different perspectives of view are shown in Visualization 1 .
Moreover, three different depth slices are also demonstrated in the imaging capability of our method, and Fig. 3(d) shows the recovered absorption images. The central slice can provide the best absorption contrast, but the absorption information vanished at defocused reconstruction planes because the observed object is a thin layer on the in-focus plane. These results together with the corresponding line profiles shown in Fig. 3(d) confirm the achievable lateral resolution (Group 11, horizontal Element 4, indicated by the white arrows). We also evaluated the depth sectioning capability of the reIDT technique by providing axial sections at different planes, as shown in Figs. 3(e) and 3(f). The object information is completely indistinguishable when the defocus distance is 0.6 μm and the FWHM of the cross-section intensity profile plot is about 1.2 μm in Fig. 3(e6), confirming a depth resolution within the 1.2 μm range (in accordance with the theoretical prediction 0.95 μm). In addition, we provide more axial sections at different planes, as shown in Figs. 3(e1)–3(e5). Figure 3(f) and supplementary
4.2 B. RI Tomography on HeLa Cell
To illustrate the biological applications of reIDT, we imaged unstained HeLa cells and achieved high-resolution RI tomography over an FOV of . Figure 4(a) shows the quantitative RI map of HeLa cells at the central plane in the reconstruction volume with 21 slices. Depth-coded projections of our reconstructions are also illustrated in Fig. 4(b) where the volumetric RI distribution is shown as a color-coded depth projection. The cell boundary and body are displayed in blue and green while the nuclear envelope and nucleolus are shown in hot colors. As shown in Figs. 4(c)–4(e), three subregions show high-resolution filopodia and other subcellular features are recovered without resolution loss. We observe cellular membrane folds, cell boundaries, microtubules, and intracellular features with high resolution [Figs. 4(c)–4(e), circles]. The nucleoli [Figs. 4(c)–4(e), arrows] inside the cell have higher RI value compared to the average index of cytoplasm, and the rendered 3D RI image provides much clearer information about the HeLa cell structure. The results shown in Figs. 4(c)–4(e) reveal that we can clearly image the details of the 3D structures of a single cell throughout its entire volume and can quantify the RI at subcellular levels. The full HeLa cell reconstruction is shown in

Fig. 4. Single-cell RI tomography of unstained HeLa cell clusters. (a) Recovered RI slice located at 0.4 μm Visualization 2 . (b) Depth color coding of 3D RI measurements of the sample in the field of view. (c)–(e) Reconstructed square subregions RI cross sections demonstrate the sectioning capability. Cellular membrane folds, cell boundaries, microtubules, and intracellular features are distinguishable (indicated by the white circles). The outlines of the nuclear envelope and nucleolus are recovered across multiple axial slices (indicated by the white arrows).
4.3 C. Tomographic Characterization of HT29
We also demonstrated reIDT’s high-resolution volumetric imaging capabilities on human colon adenocarcinoma cells (HT29). Within the RI image of FOV at 0 μm plane in Fig. 5(a), fine structures are recovered and could include microfilaments, large vacuolated mitochondria, dark granules, and lipid droplets. These dark granules can also be distinguished in the absorption slice, as shown in Fig. 5(b). We expand two regions of the reconstructed RI in Figs. 5(c) and 5(d) highlighting our depth-sectioned and high-resolution reconstructions. Dark granules ultrastructural features reported for HT29 cells include microvilli, large vacuolated mitochondria with dark granules, smooth and rough endoplasmic reticulum with free ribosomes, lipid droplets, and few primary and many secondary lysosomes [51,52]. Line profiles across multiple reconstructed slices in Figs. 5(c) and 5(d) demonstrate near-diffraction-limited lateral resolution of 346 nm. As clearly illustrated in

Fig. 5. Tomographic characterization of unstained HT29. (a),(b) RI and absorption reconstruction of HT29 in Visualization 3 .
5. CONCLUSION AND DISCUSSION
In summary, we demonstrated a label-free and scan-free intensity-only resolution-enhanced quantitative imaging method on an LED array microscope. This technique is particularly attractive for commercial microscopy, since it can achieve rapid scanning of angles by LED array illumination. By using a condenser lens, the illumination angle and efficiency are significantly increased to enable improved data acquisition. We demonstrated the technique on various samples with complex RI distributions, including the USAF target and human cancer cells, and performed 3D absorption reconstructions and quantitative characterizations of biological samples. We believe this method will set an excellent foundation for other research projects and applications, and the reIDT has the potential as a tool of great biological interest by showing its use in monitoring cell morphology and dynamics in noninvasive fast measurements.
This reIDT setup still suffers from some limitations. The 3D volume’s maximum axial resolution is limited to the objective’s DOF and can be hindered by misalignments in the LED array. Moreover, reIDT’s setup exhibits resolution limits below the diffraction limit due to mismatches between the annular array and the condenser lens. Other artifact-inducing features, such as optical aberrations and source coherence, could also be reducing the resolution limits. These factors will be evaluated in future work to provide a more accurate imaging setup. In addition, the reconstruction may be improved by more advanced algorithms, such as multiple scattering models [37,43,44] and deep learning-based 3D tomography approaches [53–58].
Article Outline
Jiaji Li, Alex Matlock, Yunzhe Li, Qian Chen, Lei Tian, Chao Zuo. Resolution-enhanced intensity diffraction tomography in high numerical aperture label-free microscopy[J]. Photonics Research, 2020, 8(12): 12001818.





