Regeneration is widely recognized as one of the most intriguing and fascinating topics in the biomedical fields[1,2]. Revealing the nature of regeneration in organisms opens a new avenue for the longstanding question of regeneration mechanisms, which provides assistance for devising therapeutic applications for humans. Recently, a rapidly growing interest in the zebrafish, which serve as a model organism of vertebrate biology for the investigation of regeneration, has appeared largely due to its pronounced regenerative capacity in several organs and tissues including the muscle, heart, pancreas, liver, skin, pigment cells, fins, and the central nervous system (CNS)[24" target="_self" style="display: inline;">–4]. More importantly, the adult zebrafish has manifested its unique ability during recovery from CNS injuries by generating new neurons to replenish the lost neural tissues[5,6].
In vivo imaging is a powerful tool for studying brain regeneration in CNS, which has the ability to characterize the damaged tissue, measure the safety and efficacy of therapy, and monitor the regeneration process. In particular, the neuroimaging techniques including magnetic resonance microscopy and ultrasound microscopy enable us to noninvasively measure the changes of the brain structures and functions based on the adult zebrafish model[7,8]. However, the imaging resolution (50–100 μm) and contrast as well as the imaging speed from the two methods mentioned above are insufficient for identifying the volume changes of brain structures in real time. In contrast, optical imaging methods including confocal and two-photon microscopy have shown their merits in capturing the structures of the zebrafish brain at embryonic stages with a high resolution (0.5–5 μm)[9–12" target="_self" style="display: inline;">–12]. However, the zebrafish brains will lose their transparency after their first two-weeks of development, and most available optical imaging methods will not be able to effectively image the zebrafish brain at the adult stage. As such, it is crucial to develop and use new optical imaging techniques which can achieve high-resolution imaging of the adult zebrafish brain with an excellent penetration depth and video-rate imaging speed.
Optical coherence tomography (OCT) is a robust and attractive imaging method that uses scattering light to reconstruct the three-dimensional (3D) images of biological tissues with a micrometer resolution[1315" target="_self" style="display: inline;">–15]. The most recent advance in OCT technologies is the development of the spectral-domain (SD) OCT imaging system, which has shown promise in the biological and biomedical imaging field[16–19" target="_self" style="display: inline;">–19]. Compared to other approaches used for imaging the zebrafish brain, OCT has competitive advantages in that both the image quality and the imaging depth is effective for providing detailed 3D information on the micro-structure of an intact adult zebrafish brain. In this study, a long-range SD-OCT system was employed for the long-term monitoring of brain tissue regeneration using an adult zebrafish model of brain injury. It is suggested that the long-range SD-OCT system used in this study is able to provide high-resolution and real-time monitoring of brain regeneration at different stages in a mechanically damaged brain of an adult zebrafish.
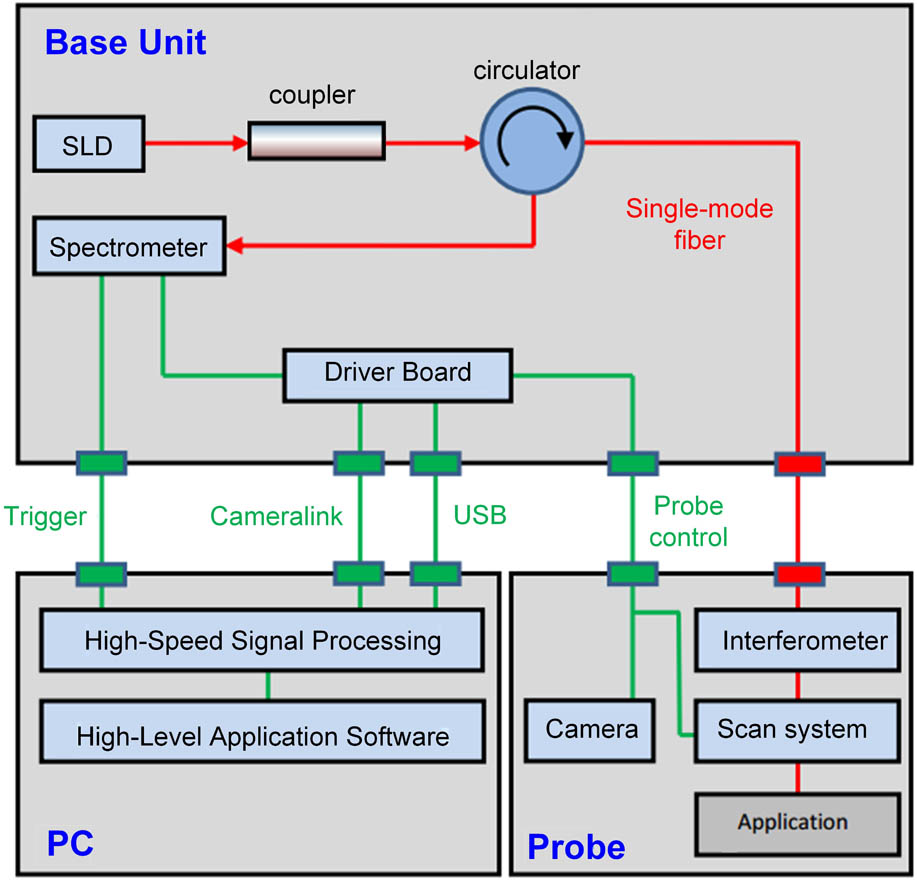
Figure 1 displays the standard Thorlabs SD-OCT (TELESTO-II-1325LR) imaging system that was utilized for the present work. This SD-OCT system applies a single super luminescent diode (SLD) with a 1325 nm central wavelength and an over 100 nm spectral bandwidth as the light source. The spectrometer design is optimized for the light source of this system, and our SD-OCT imaging setup can achieve an axial resolution of 12 μm and a lateral resolution of 13 μm in the air. The imaging depth of this system can achieve around 6 mm for the agar phantom and about 2 mm for tissues such as chicken breast[20]. The scan system adopts two high-speed galvo mirrors, which enables rapid volume acquisitions at high imaging speeds of up to a 76 kHz A-scan per second. As a result, the SD-OCT can achieve real-time, depth-resolved, cross-sectional, and 3D imaging. The maximum viewing field of the SD-OCT system can reach . Data acquisition and signal processing were simultaneously performed using a high-performance personal computer. A CCD camera could lively monitor the application area and take photographs of the samples. OCT imaging will be performed along a path that is selected on the photograph of a sample. With the help of the CCD camera, we are able to acquire the OCT images along the same profile on the zebrafish head for each time point.
Fig. 1. Schematic of our SD-OCT system for imaging the injured brain of the adult zebrafish.
下载图片 查看所有图片
Transparent casper zebrafish (male, 120 days) were used in the present study. Zebrafish were maintained in our flow-through aquaria at a temperature of with the photo period of 14 h light and 10 h dark[21]. All procedures were performed in compliance with the guidelines on animal research stipulated by the Animal Care and Use Committee at the University of Macau. Before the brain injury, the adult zebrafish were first anesthetized with 0.01% MS-222 in system water, and then placed under the microscope of our SD-OCT system. Guided by the SD-OCT imaging, a sterile 25-gauge needle was inserted into the head to induce the brain injury. Immediately after the brain lesion was developed, the animals were put back into the fresh water. After the brain injury, our SD-OCT system was used to monitor the tissue regeneration of the mechanically damaged brain. The long-term monitoring and imaging of the recovery of the brain injury were performed at days 0, 10, 20 and 43 after the brain lesion was induced. The zebrafish were divided into the control group and brain injury group with three zebrafish in each group. Before the brain regeneration experiment, a histological analysis was conducted to characterize the healthy brain and injured brain by sacrificing one zebrafish from each of the two groups.
For the SD-OCT imaging at each time point, the fish were first anesthetized using 0.01% MS-222 in system water until they became unresponsive to the touch[20]. Then the fishes were moved to a petri dish containing two plexiglass panels that could keep the fish’s body upright. Further, the fish were placed in the field of view under the scan objective lens. After adjusting the focal plane to the suitable position upon the head of the fish, we performed both the cross-sectional and 3D imaging of the adult zebrafish brain at the video-rate imaging speed. Again, once the experiment was completed at each time point, the zebrafish were put back into the fresh water. Finally, the data was processed to generate several sets of high-resolution 3D brain images at different time points.
A histological experiment to characterize the normal and injured brain was performed by sacrificing the zebrafish. Zebrafish embedded inside paraffin sections were cut at a thickness of 100 μm, and stained with hematoxylin and eosin (H&E). Images were generated using a Nikon microscopy with a 2 times objective, and the imaging results were given in Fig. 2. In particular, Fig. 2(a) showed the H&E staining of the normal head, from which we can see that the normal brain tissue has the symmetrical structures and was covered with an intact skull bone. In contrast, as plotted in Fig. 2(b), a significant lesion located on both the skull and the brain were clearly identified for the injured head, which was labeled with green dash curve. It was expected that the lesion was filled with thrombus that was quickly accumulated to stop the bleeding after the mechanical injury.
Fig. 2. Histological (H&E) study of adult zebrafish head (a) without or (b) with the needle insertion induced brain injury. Both images were generated using a Nikon microscope under a 2 times objective.
下载图片 查看所有图片
After brain injury was induced in the adult zebrafish using a 25-gauge needle (0.5 mm in diameter), SD-OCT imaging was implemented to capture the location and size of the resultant lesion. It was observed in Fig. 3 that the injured areas were clearly identified by the SD-OCT imaging system at 0 days after we inserted the needle into the adult zebrafish brain. We first took the representative slices from the generated images for the analysis of the skull and brain injury due to the inserted needle. We found in Fig. 3(a) that the damaged area of zebrafish skull was clearly localized, as shown in the semicircle groove region labeled with the blue dash curve. In addition, one sagittal and one coronal slice, as displayed in Figs. 3(b) and 3(c), were provided to further reveal the detected brain injury lesion and the damaged skull by using our 3D SD-OCT system. We can observe in Fig. 3(b) that significant skull bone damage was identified in the region circled by the blue curves. In addition, a brain lesion with low imaging intensity (labeled with the green dash curve) was detected beneath the damaged skull, as shown in Figs. 3(b) and 3(c). The coronal and sagittal images enable a depth measurement of the lesion inside the brain tissue (). The low intensity region was generated by the loss of brain tissues due to the insertion of the needle, which should be filled with lipid or fluid. In particular, the zebrafish brain owns the trait of a high level of bilateral symmetry, which was actually altered after the brain injury at 0 days post-lesion, as displayed in Fig. 3. It is noted from the reconstructed results in Fig. 3 that the inserted needle could induce injury to both the skull and the brain of an adult zebrafish, which was able to be detected by our 3D SD-OCT imaging system.
Fig. 3. High-resolution SD-OCT imaging of the needle insertion induced brain injury. (a) Visualization of the needle insertion induced skull injury reconstructed by the 3D SD-OCT. The blue curve denotes the location of the injured regions. (b) The coronal SD-OCT image of the brain along the solid red curve at top right corner across the lesion. The green dash curve denotes the location of the lesion. (c) The sagittal SD-OCT image of the brain lesion along the red line at the top right corner. (d) The sagittal SD-OCT image of the intact brain along the red line at the top right corner. The scale bar is 250 μm.
下载图片 查看所有图片
Unlike mammals, zebrafish exhibit extensive neural regeneration after brain injury in the adult stages of its lifetime. In the study of brain regeneration, 3D in vivo imaging is indispensable for identifying the structural changes of damaged tissues, and for measuring the safety and efficacy of the treatment of biological systems. To monitor and detect the process of tissue regeneration, we performed the 3D in vivo imaging in a zebrafish model of adult brain injury using our SD-OCT system during a period of 43 days. The coronal and sagittal views of the injured zebrafish brain at each time point (0, 10, 20, and 43 days post-lesion) were provided to show the changes of the brain lesion in detail, as plotted in Figs. 4(a) (four coronal slice images) and 4(b) (four sagittal slice images). The SD-OCT images for an injured zebrafish brain at 0 days post-lesion were given at the top left of Figs. 4(a) and 4(b), where we can see that the brain lesion and the loss of brain tissues (labeled with the green dash curve) as well as the loss of the skull bone (labeled with the blue dash curve) were clearly identified by the SD-OCT. Here a sharp edge labeled with a green curve in the sagittal SD-OCT image [top left of Fig. 4(b)] was used to separate the injured and intact regions of the brain. The injured areas might initially be filled with fluid and lipids that generally had low optical scattering coefficients. Interestingly, until 10 days post-lesion, new tissues were beginning to accumulate in the traumatic brain injury regions, and the brain lesion size was also reduced significantly, as shown at the top right of Figs. 4(a) and 4(b). At 20 days post-lesion, considerable recovery was observed for the skull bone, as shown at the bottom left of Figs. 4(a) and 4(b). Likewise, the brain lesion also gradually repaired itself, and the symmetrical structures of the adult zebrafish brain began to occur again only 20 days after the traumatic brain injury. Further, when compared to the sagittal SD-OCT image with the intact brain in Fig. 4(d), we found the brain regeneration still continued until the brain totally repaired itself at 43 days post-lesion. More importantly, new tissues were generated between the fully recovered skull and brain tissues [labeled with red arrow at the bottom right of Fig. 4(a)], which might be the hyperplasia from skull bone. Our findings suggested that adult zebrafish have an extraordinary capability in neural regeneration after the loss of the brain tissues.
Fig. 4. Time Serial SD-OCT images show the injured adult zebrafish brain at 0, 10, 20, and 43 days post-lesion. (a) Coronal SD-OCT views of the brain lesion at different time points after the brain injury. (b) Sagittal SD-OCT view of the brain lesion at different time points after the brain injury. The dashed blue curve indicates the injured skull regions, whereas the dashed green curve denotes the approximated boundary of the zebrafish brain.
下载图片 查看所有图片
A more direct characterization of brain tissue regeneration is enabled by 3D visualization of the whole brain. Figures 5(a) and 5(b) showed the 3D SD-OCT images of the adult zebrafish brain in volumetric and cross-sectional form, respectively. The imaging results, as shown in Figs. 5(a) and 5(b), indicated that the SD-OCT was able to map the whole head of the adult zebrafish. As we can see, the zebrafish brain was covered by the skull and other bones, and in particular, some bones are even shown to have the structures of honeycomb (labeled with the red dash curve). To view the internal structures of the zebrafish brain, the 3D SD-OCT images could be viewed in terms of three orthogonal planes, as shown in Fig. 5(b), in which the ventricle was captured and located at the tectum optimum area (labeled with blue dash curve). Furthermore, the 3D OCT image could be continuously segmented, so that the planes of any orientation can be clipped to expose structures within the volume. The segmented results of the 3D OCT image of the intact adult zebrafish brain were presented in Video 1.
Fig. 5. 3D characterization of the intact brain as well as the brain tissue regeneration process. (a) The 3D view of the intact adult zebrafish brain (volumetric form, Video 1). (b) The 3D view of the intact adult zebrafish brain (cross-sectional form, Video 2). (c) The cross-sectional 3D view of the brain lesion at 0 days post-lesion (Video 3). (d) The cross-sectional 3D view of the brain lesion at 43 days post-lesion (Video 4).
下载图片 查看所有图片
The 3D SD-OCT images for an injured zebrafish brain were reconstructed at days 0 and 43 post-lesion. Figure 5(c) showed the cross-sectional 3D view of the brain lesion at 0 days post-lesion in which the loss of brain tissues and skull bone (labeled with green dash curve for the X-Z slice and red dash curve for the X-Y slice) were clearly observed. Again the low imaging intensity in the lesion regions is due to the low optical scattering coefficient of the fluid tissues distributed along the injured brain areas. In addition, Fig. 5(d) showed the cross-sectional view of the brain injury region at 43 days post-lesion in which the low scattering areas as well as the brain lesion areas totally disappeared. Based on the brain mapping results of the adult zebrafish by the 3D SD-OCT, it is found that the zebrafish can repair itself after a brain injury.
In this study, long-term in vivo monitoring of brain regeneration of the adult zebrafish is implemented from the day of injury to 43 days of recovery by using our SD-OCT system. SD-OCT imaging is able to noninvasively visualize the skull injury and brain lesion of the adult zebrafish in real time with a high spatial resolution.
Jian Zhang, Zhi-Wei Zhang, Wei Ge, Zhen Yuan. Long-term in vivo monitoring of injury induced brain regeneration of the adult zebrafish by using spectral domain optical coherence tomography[J]. Chinese Optics Letters, 2016, 14(8): 081702.