The organization of neurobiological tissue containing the arrangement of cells or the relative amount of cell numbers in some special layers and brain regions is crucial to our understanding of the structure and function of the brain[1–4" target="_self" style="display: inline;">–4]. Magnetic resonance imaging (MRI) and positron emission tomography (PET) have been employed into brain structure and function research, but the relatively low spatial resolution and tissue contrast hamper them in providing organization morphology with a resolution enough to distinguish cells[5,6]. Fluorescence microscopy is powerful in the research of the brain structure and function with submicron spatial resolution[7,8], while it often needs labeling fluorescent indicators, such as fluorescent proteins and chemical dyes, to provide an enough contrast. Coherent anti-Stokes Raman scattering (CARS) microscopy, based on the molecular vibration in the tissue, can achieve label-free biochemical imaging with high spatial resolution, but the relative low sensitivity and poor contrast of detection, due to an unavoidable lower resonant background[9,10], restrict its application to brain study.
Second harmonic generation (SHG) microscopy, based on the nonlinear interaction mechanisms, not only has the capability of submicron resolution and inherent optical sectioning, like two-photon excitation fluorescence (TPEF) microscopy[11,12], but also provides label-free imaging for some structural proteins, such as collagen, actomyosin, and tubulin[13]. Although the extracellular matrix in brain tissue is short of collagen, SHG microscopy has succeeded in imaging a label-free acute hippocampal slice and cultured living neuronal cells, which have an abundance of rich uniform polarity microtubules in their axon structure[1416" target="_self" style="display: inline;">–16]. Combined some special dyes, SHG microscopy also serves to record the electrical activity in intact neuronal networks[1719" target="_self" style="display: inline;">–19]. However, at present, there has no study to report on the structure and organization of the cerebellum and brain stem using the label-free SHG imaging technique. Recently, studies show that, besides the major role in motor function, the cerebellum might contribute to diverse aspects of behavior[20,21]. The brain stem controls the flow of messages between the brain and the rest of the body, and it controls basic body functions, such as breathing, swallowing, heart rate, blood pressure, and consciousness[22]. Therefore, we now try to study the cerebellum and brain stem using SHG microscopy.
In this Letter, by using a custom-built large area SHG microscope, we first demonstrate it to characterize the morphological features of the intact coronal brain slice of the cerebellum and brain stem. From the SHG images, different brain regions and subregions can be easily distinguished from their morphology, and the detailed structures can be depicted. Moreover, due to cell soma appearing as dark shadows on the SHG image, we can easily extract the cell number in each layer of cerebellum. Interestingly, on the SHG image the soma and axon initial segment (AIS) structure of a Purkinje cell (PC) can be easily recognized as a brush surrounded by the SHG signals, maybe from axons of basket cells. Our results demonstrate that the custom-built large area SHG microscopy provides a useful tool for the research of brain organization and structure.
In this study, the GAD67-GFP adult transgenic female mice were used. The Animal Experimentation Ethics Committee of Huazhong University of Science and Technology approved animal care and experiments protocols. The mouse was anesthetized with 0.4–0.6 mL mixture containing 0.625 mg/mL xylazine and 6.25 mg/mL ketamine. After being decapitated, the whole brain was dissected and sectioned into 300 μm coronal slices using a vibrating microtome (VT1000S, Leica). The brain slices were fixed in 4% paraformaldehyde for 24 h and, then, transferred into phosphate buffer solution (PBS) for SHG imaging.
Imaging was performed on a custom-built large area SHG microscope[16]. A Ti:Sapphire laser (Maitai BB, Spectra-Physics), working at wavelength of 760 nm, was scanned by a galvanometer mirror scanning system (Model 6215, Cambridge Technology). An apochromatic objective (UPLSAPO , Olympus) was employed to focus the excitation beam and collect the emission signals. Through a band-pass filter (380/14 nm, Semrock), SHG signals were detected by a photomultiplier tube (PMT, H7422A-40, Hamamastu). In addition, to co-locate SHG images, green fluorescent protein (GFP) fluorescence signals were also collected. The fluorescent signals were split out by a dichroic mirror (FF409, Semrock) and detected after a band-pass filter (525/40 nm, Semrock) by the other PMT (H7421-40, Hamamastu). The dwell time per pixel was 24 μs and each image () took . A translation stage (ES111, Prior Scientific) was used to translate the specimen after an image was acquired. Finally, sequence images were combined into an intact brain slice image with a size of composed of images of . A Nikon upright A1R microscope (Minato, Tokyo, Japan) with a water immersion objective (Nikon, Minato, Tokyo, Japan) was used to detect the spectrum.
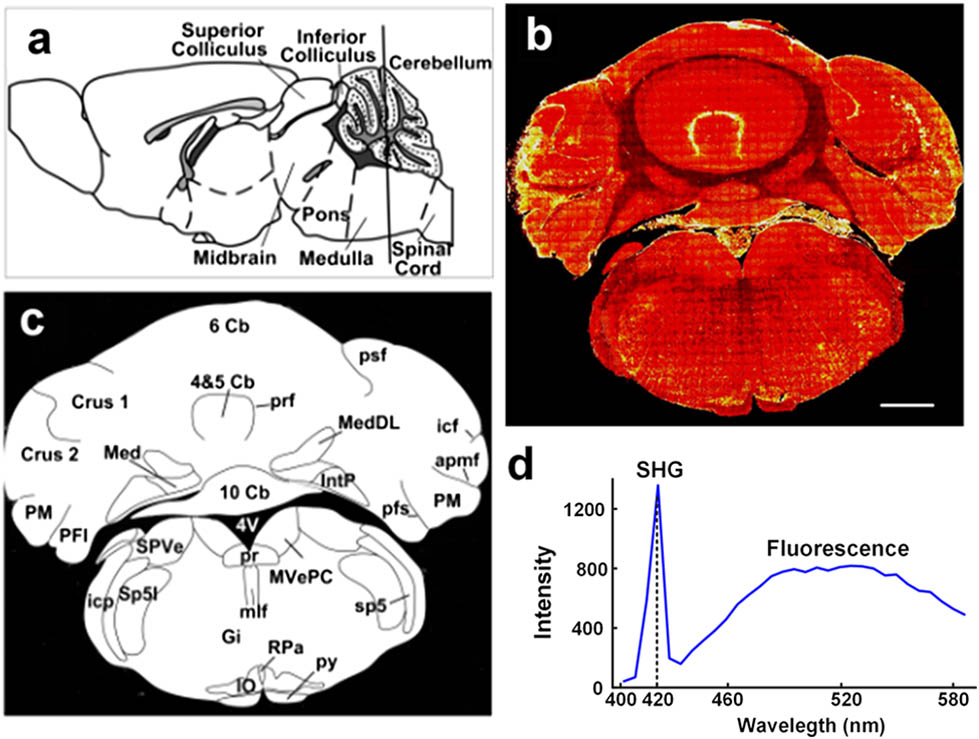
Modern neuroscience is increasingly macroscopic in characterizing and discriminating different brain regions as a means for understanding complex brain anatomy and localizing experimental data. An SHG image of the brain slice is shown in Fig. 1. Figure 1(a) indicates the schematic position of the brain slice imaged in this study. In Fig. 1(b), the contours and subregions can be seen from the SHG image of a coronal cerebellum and brain stem slice. Figure 1(c) is the corresponding coronal map of the brain slice in Fig. 1(b). From a comparison of Figs. 1(b) and 1(c), the macroscopical features of different regions, such as cerebellar lobules, cerebellar nucleus, inferior olive (IO), pyramidal tract, and so on, are clearly observed. These regions can be easily discriminated, according to the tissue morphology in Fig. 1(b): fourth and fifth cerebellar lobules (4 and 5 Cb); sixth cerebellar lobules (6 Cb); 10th cerebellar lobules (10 Cb); crus 1 of the ansiform lobule (Crus1); crus 2 of the ansiform lobule (Crus 2); paramedian lobule (PM); paraflocculus (PFl); posterior part of interposed cerebellar nucleus (IntP); medial cerebellar nucleus (Med); dorsolateral protuberance of the medial cerebellar nucleus (MedDL); primary fissure (prf); posterior superior fissure (psf); intercrural fissure (icf); ansoparamedian fissure (apmf); parafloccular sulcus (pfs); spinal vestibular nucleus (SPVe); magnocellular part of medial vestibular nucleus (MvePC); interpolar part of spinal trigeminal nucleus (SP5I); prepositus nucleus (Pr); medial longitudinal fasciculus (mlf); gigantocellular reticular nucleus (Gi); raphe pallidus nucleus (Rpa); IO; pyramidal tract (py); fourth ventricle (4V). The structures of the intact brain slice revealed by SHG imaging are consistent with the anatomical structures previously reported[17].
Fig. 1. From an SHG image to the structure of brain slice. (a) The position schematic of the coronal brain slice. (b) Morphology of a cerebellum and brain stem slice detected from SHG imaging. (c) Schematic of the discernable regions in SHG image (b). (d) The spectra of the slice with the Nikon A1 Microscope using an 840 nm laser. Scale bar in (b) is 1 mm.
下载图片 查看所有图片
To further clarify these characteristic signals on our SHG image, we tried to provide spectroscopic evidence using a commercial microscope. Here, the Nikon upright A1R microscope was used to detect the spectrum. For a wavelength detection range above 400 nm, we employ an 840 nm laser, and an emission intensity of range 402–594 nm was detected with 32 channels. The spectroscopic curve is shown in Fig. 1(d); from the image, it is obvious that there is a narrow emission at 420 nm (just half of 840 nm) and a following wide emission (maybe the autofluorescence), with distinct waveforms. So, it suggests that the image of Fig. 1(b) is from SHG signals.
Further, combining with TPEF microscopy and transgenetic mouse GAD67, we tried to demonstrate the SHG microscope to image the detailed morphological feature of the cerebellum and brain stem slice, containing information of the cell’s arrangement and architecture. Except for SHG images, the corresponding fluorescence images of the tissues were simultaneously acquired with a PMT (H7421-40, Hamamastu) after a band-pass filter (525/40 nm, Semrock). In Fig. 2(a), molecular layer (ML), PC layer (PCL), and granule layer (GL) of the cerebellar cortex can be easily identified. The black, blue, and white arrows indicate the neuron somas, which cannot produce SHG due to the poor phase matching. The TPEF image of cerebellar cortex is shown in Fig. 2(b). In the TPEF image, the PCs and interneurons (basket and stellate cells) are visualized by high fluorescence intensity, owing to GFP expression in the neuron somas, while granule cells can also be located by rare fluorescence for no GFP expression[7]. The image combining SHG and TPEF signals is shown in Fig. 2(c). The result indicates that the SHG and TPEF signals are complementary at PCs and interneurons somas, but provide identical structure features at the GL. For the other cerebellar regions and brain stem regions that contain plentiful neurons, the characteristic of SHG imaging is similar to the results from the cerebellar cortex. In addition, the images of white matter (WM) in the cerebellum, inferior cerebellar peduncle (icp), and spinal trigeminal tract (SP5) regions, which are rich in nerve fibers, are also shown in Fig. 2. The results prove that SHG signals can provide consistent morphological features with TPEF image for nerve fibers.
Fig. 2. (Color online) Combined SHG and TPEF images of the mouse cerebellum and brain stem slices. (a) SHG image shown in hot colormap, (b) TPEF image in green, (c) the merged SHG/TPEF image of the cerebellar cortex. The black, blue, and white arrows indicate the neuron somas in ML, PCL, and GL, respectively. (d)–(f) are the SHG image (hot colormap), TPEF (green), and the merged images SHG/TPEF of icp and SP5. Scale bars in (c) and (f) are 100 μm.
下载图片 查看所有图片
Detailed structures of different brain regions are shown in Fig. 3. The MvePC, SpVe, Gi, IO, mlf, and py regions of the brain stem are shown in Figs. 3(a)–3(f) with the cell somas indicated by blue arrows. In particular, the white arrows in Figs. 3(b) and 3(c) show the reticular formations in the SpVe and Gi regions[23]. In addition, Figs. 3(e) and 3(f) reveal that the mlf and py regions are rich in nerve fibers instead of neuron somas[24,25]. A cerebellar fissure is shown in Fig. 3(g), which divides the cerebellum into small lobes. The white and green arrows in Fig. 3(h) indicate the blood vessel and apical dendrites, respectively. SHG images of deep cerebellar nucleus (MedDL, Med, and IntP) are shown in Figs. 3(i)–3(k). Different from the Med and IntP region, the MedDL region is rich in neurons in the section.
Fig. 3. (Color online) Detailed structures of different brain stem and cerebellar regions obtained by label-free SHG imaging. (a)–(f) the images of the MvePC, SpVe, Gi, IO, mlf, and py regions. Blue and white arrows indicate cell somas and the reticular formations, respectively. (g) Cerebellar fissure indicated by the white arrow. (h) Blood vessel and apical dendrites are marked by white and green arrows, respectively. (i)–(k) Deep cerebellar nucleus images of MedDL, Med, and IntP regions. Blue arrows indicate cell somas. (l) Magnified image of PCL. Blue and white arrows indicate PCs and the AIS structure of the PC, respectively. Images shown in hot colormap. Scale bar is 50 μm.
下载图片 查看所有图片
Cell density is another subject of major concern for neuroscientists. It has been reported that many neurons are susceptible to many exposures, such as alcohol or lithium and autoimmune diseases[2628" target="_self" style="display: inline;">–28]. For example, the number of PCs would decrease in the autistic brain[28,29]. Compared to normal people, the mean number of Purkinje neurons was also 20% lower in the cerebellum of schizophrenia and bipolar disorder patients[30]. Therefore, it is useful to estimate the cell number in different layers and regions of the brain for physiological and pathological research.
In this study, the cell density was calculated according to the following steps: select multiple areas () with the same size in the brain region of interest, then manually count the cell number and calculate the cell density within the areas; calculate the mean density of multiple areas and express as deviation (SD). Table 1 shows the approximate densities of cells in different cerebellar cortex regions and the deep cerebellar nucleus region. As opposed to the low density of cells in the ML, PCL, and cerebellar nucleus, small granule cells are densely packed in the GL[31]. These results suggest that SHG microscopy can not only provide the morphological features of the cerebellum and brain stem, but also quantify the density of the cells in the cerebellum to study functions of the brain in future research.
Table 1. Densities of the Cells in Different Layers and Regions of the Cerebellar Cortex.
| Layers and Regions of the Cerebellar | Density of the Cells () |
|---|
| ML | | | PCL | 31.4 ± 15.1a | | GL | | | Deep cerebellar nucleus | |
|
查看所有表
It is mentioned that SHG processes require noncentrosymmetric proteins, like collagen and microtubule[14,32]. Witte et al. have tried SHG in addition to THG imaging, but found that the SHG signal of gray matter is too weak to be detected due to the absence of collagen in the brain’s extracellular matrix and its amorphous structure[33]. In this study, a high quality SHG image of the cerebellum and brain stem was acquired. The reasons may be as follows: firstly, nondescanned detection, the most efficient fluorescence collection scheme, and a GaAsP photocathode PMT with high quantum efficiency ( QE at peaking wavelength) used in the homebuilt system can provide high SHG signals-detection efficiency. Secondly, it might be noted that, although SHG signals initially propagate with the incident laser in the forward direction in the coherent process, the back scattering SHG signals are observed throughout the brain slice due to the multiple scattering events arisen from the small scatters in brain tissue with a size less than an SHG wavelength[34]. Lastly, although it is absent of collagen in the brain’s extracellular matrix and its amorphous structure, the cerebellum and brain stem tissue has plenty of enriched microtubule axon fibers, through which it is connected with other brain regions. Perhaps, this characteristic makes the cerebellum and brain stem easier to image than other brain regions.
In addition, just due to the enwrapping by the exuberant terminal branches of basket cells axon, the fine structure of the AIS of PCs can be recognized on the SHG images. As shown in Fig. 3(l) (indicated by white arrows), the AIS of PCs can be precisely located. In addition, it is consistent with the fine organization of the cerebellar pinceau-basket axon collaterals extending around the Purkinje soma, synapsing on the Purkinje AIS, and forming the pinceau[35,36]. Since the AIS of the PC is enwrapped by the exuberant terminal branches of basket cells axon[7,35], the strong label-free SHG signals from the axons of basket cells make the soma and AISs of PCs visible. This SHG-based imaging would also be used to guide micropipettes toward designated neurons in live tissue for electrophysiology.
In conclusion, we achieve label-free monitoring the organization and structures in different layers and regions of a mouse cerebellum and brain stem slices by a homebuilt large area SHG microscope. In addition, we evaluate the cell densities in different layers and regions dedicated to a particular modality based on SHG images. Combining morphological analysis and quantitative analysis of the tissue, our results demonstrate that the large area and label-free SHG microscopy is helpful for structural and functional brain research and has the potential to further benefit the neuropathic disease study.
Xiuli Liu, Daozhu Hua, Ling Fu, Shaoqun Zeng. Imaging the structure and organization of mouse cerebellum and brain stem with second harmonic generation microscopy[J]. Chinese Optics Letters, 2017, 15(9): 090003.
 Download: 1113次
Download: 1113次