浮游藻类细胞显微明场图像与荧光同步测量图像配准方法研究  下载: 616次
下载: 616次
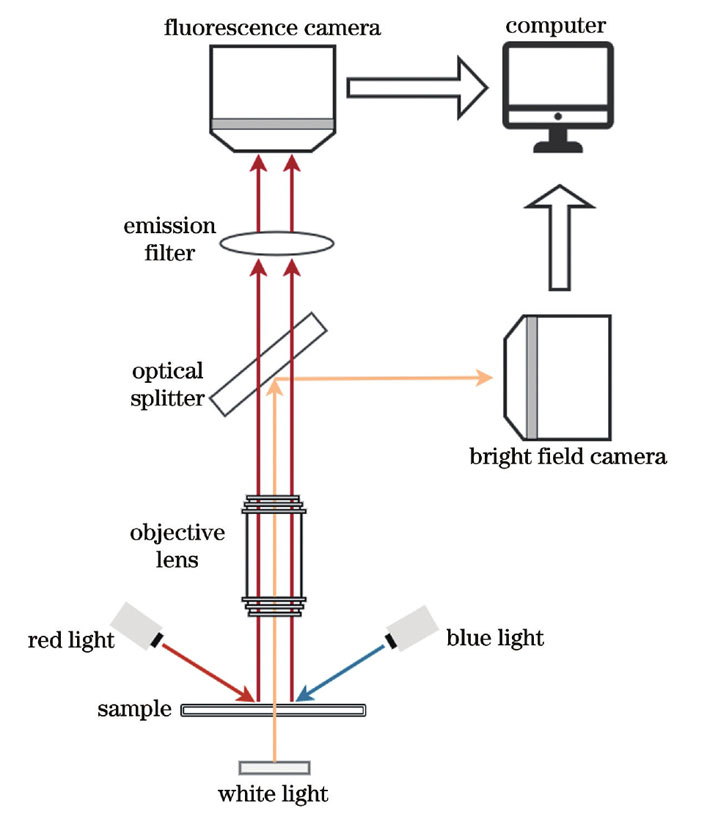
Detection of phytoplankton diversity is an important part of a water quality bioassessment. The traditional manual microscopic detection of algae species requires professional operation and is time-consuming and laborious; therefore, these challenges can be overcome by the development of a method for automatic identification of phytoplankton cell images. Similar to manual identification, deep learning and other automatic identification technologies identify phytoplankton cells based on the morphological characteristics of bright field cell images; however, the practical applications of such technologies encounter complications, such as the difficulty of accurate segmentation of phytoplankton cells and the limitation in high recognition accuracy of algae species only with a small range of groups. Previous studies have shown that the accuracy of algal cell segmentation and recognition can be effectively improved by fusing the bright field and fluorescence images of phytoplankton cells. However, the fusion of bright field and fluorescence images synchronously measured from phytoplankton cells requires significantly high accuracy and shockproof capability of the mechanical structure of the acquisition system. Under a high-power microscope, even a small error in the mechanical structure or a small vibration of the camera can lead to a dislocation between the bright field and fluorescence images, thus causing difficulties in the fusion of the images. Therefore, the registration of bright field and fluorescence images of algae is of great significance for the automatic identification of phytoplankton.
The displacement between bright field and fluorescence images can be represented by a rigid transformation model, which consists of three parameters: the translation in the x direction, translation in the y direction, and rotation angle. Normalized mutual information is used to calculate the similarity between the bright field and fluorescence images. The goal of image registration is to find a set of rigid transformation parameters that maximize the normalized mutual information of the two images. Owing to the significant difference between the bright field and fluorescence images of phytoplankton cells, the similarity is difficult to be characterized by directly calculating the normalized mutual information. In this study, the normalized mutual information of the S channel binary image of the bright field HSV color space and the binary image of the fluorescence gray level was assumed as the similarity between the bright field and fluorescence images. The S component of the bright field HSV image and the fluorescence gray image were decomposed using a two-dimensional discrete wavelet transform, and the low-frequency components were binarized, to accelerate the registration. First, the particle swarm algorithm was used to register the low-frequency components of the five-level wavelet decomposition. Subsequently, the translation and rotation angle of the preliminary registration were assumed as the initial values, and the low-frequency components of the wavelet three-level decomposition are further registered using Powell’s algorithm.
Scenedesmus sp., Selenastrum capricornutum, and Nostoc sp. are used as experimental objects. The similarity and registration methods are compared and analyzed. As shown in Figs. 3 and 4, the normalized mutual information of the bright field S channel and the fluorescence grayscale image has an obvious peak value after binarization, which is extremely conducive to parameter optimization in the registration process. As shown in Fig. 5, after the bright field and fluorescence images are decomposed by the wavelet, the noise in the high-frequency component is concentrated, and the low-frequency component has a higher normalized mutual information. Therefore, the normalized mutual information of binary images of low-frequency components after wavelet decomposition is chosen as the similarity measurement index for bright field and fluorescence images. Table 1 presents an experimental comparison between the proposed method and other registration methods. Compared with particle swarm optimization algorithm, the proposed method reduces the mismatch rate by 0, 8.1, and 0.7 percentage points, shortens the running time by 128.26 s, 448.95 s and 237.20 s, and improves the normalized mutual information by 0.065, 0.083, and 0.106; Powell’s method depends on the initial value; therefore, it is easy to fall into the local maximum in the process of optimization, which leads to a high mismatch rate; compared with GA, the mismatch rates of proposed method are reduced by 6.1, 26.2, and 10.2 percentage points, the running time is shortened by 23.78 s, 60.95 s and 33.74 s, and the normalized mutual information after registration is improved by 0.149, 0.170, and 0.180. The experimental results demonstrate that the proposed method has obvious advantages in terms of registration accuracy and running time compared with other registration methods.
In this study, the bright field image is converted into a binary image by processing the S channel of the HSV color space; the fluorescence gray image is binarized; normalized mutual information is used as the similarity criterion to characterize the similarity of the bright field and fluorescence image. Using Scenedesmus sp., Selenastrum capricornutum, and Nostoc sp. as experimental objects, the application of wavelet decomposition in the registration of microscopic bright field and fluorescence images of phytoplankton cells was studied. The global search advantage of the particle swarm optimization algorithm is used for preliminary registration in the high-level decomposition component, and the local search ability of Powell algorithm is used for fine-tuning the registration accuracy in the low-level decomposition component. A comparative analysis is performed with other commonly used registration methods, and the experimental results verify the feasibility of the registration method.
贾仁庆, 殷高方, 赵南京, 徐敏, 胡翔, 黄朋, 梁天泓, 何前锋, 陈晓伟, 甘婷婷, 张小玲, 马明俊. 浮游藻类细胞显微明场图像与荧光同步测量图像配准方法研究[J]. 中国激光, 2022, 49(24): 2407202. Renqing Jia, Gaofang Yin, Nanjing Zhao, Min Xu, Xiang Hu, Peng Huang, Tianhong Liang, Qianfeng He, Xiaowei Chen, Tingting Gan, Xiaoling Zhang, Mingjun Ma. Registration Method of Microscopic Bright Field and Fluorescence Synchronous Measurement Images of Phytoplankton Cells[J]. Chinese Journal of Lasers, 2022, 49(24): 2407202.