Compact and effective photon-resolved image scanning microscope
1 Introduction
Asynchronous read-out single-photon avalanche diode (SPAD) array detectors1
We integrate our new DFD-DAQ and control system into an SP-LSM architecture equipped with a commercial SPAD array detector. We validate the DFD module embedded in the microscope control and DAQ unit by implementing different FLISM-based imaging techniques. We combine our new platform with the fluorescence lifetime phasor analysis to implement functional superresolution imaging. Despite the lower temporal precision of a typical TT-DAQ module, the DFD-DAQ module allows the investigation of timescales short enough to monitor the lifetime changes of molecules with biological interest. The combination of our architecture with the phasor analysis also enables superresolution imaging of multiple fluorophores without spectral emission separation. In this case, we distinguish the different dyes on a sample by leveraging their different fluorescence lifetimes. Furthermore, we took advantage of the integration of the DFD module into the microscope control system to implement a pulse-interleaving multiwavelength excitation scheme. Different wavelengths enable the excitation of multiple fluorophores, which might have overlapping absorption spectra. We introduce a phasor-based method for multispecies separation, also in the event of spectral excitation cross talk. Finally, we implement nanoscopy by combining STED-ISM with separation-by-lifetime tuning (SPLIT).25,26 This approach uses the phasor analysis and the fluorescence lifetime changes induced by the stimulation emission process27 to improve the resolution of STED microscopy. Our results demonstrate the versatility and vast potentialities of the proposed architecture.
2 Results
In this work, we used a custom fluorescence laser-scanning microscope [Fig. 1(a)] equipped with a SPAD array detector, a pair of triggerable picosecond pulsed diode lasers for excitation (560 and 640 nm), and a three-dimensional scanning system. A subnanosecond pulsed fiber laser (775 nm), synchronized with the excitation lasers, enables stimulated emission for STED microscopy. The FPGA board includes both the multichannel DFD-DAQ and the microscope control modules. The first records the inputs from each channel of the SPAD array detector; the latter provides the analog outputs for driving the scanner and the digital outputs for triggering the pulsed lasers. Since both modules have common clocks, their synchronization is straightforward: the lifetime histogram is calculated on the FPGA board and returned at the end of each pixel dwell time. Furthermore, the trigger signals for the lasers can be delayed to enable pulse-interleaving excitation—namely, alternating excitation at different wavelengths. For each scan point and each channel of the SPAD array, the FPGA builds the photon arrival histogram using 25 low-precision time-to-digital converters (TDCs) based on the DFD principle. The DFD system computes the photon arrival time by measuring the delay between the excitation event and the digital single-photon signal coming from the SPAD.
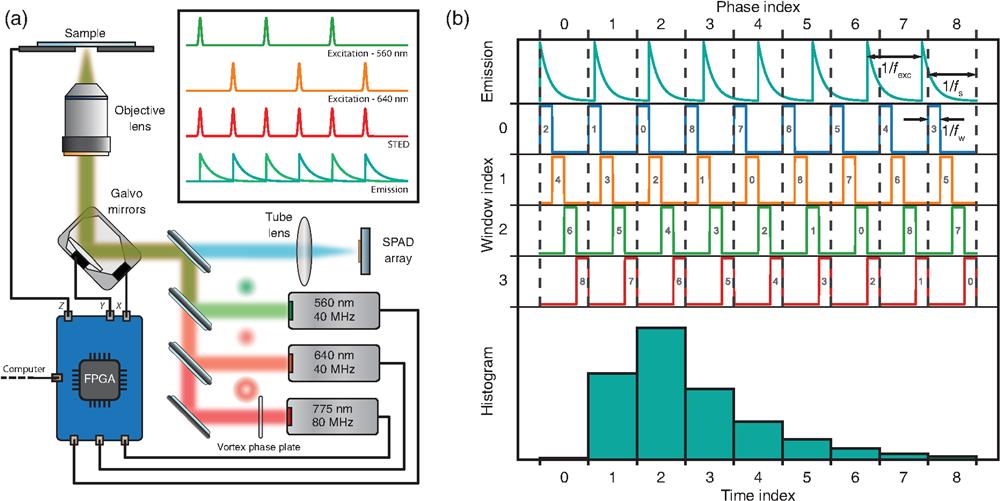
Fig. 1. Setup and acquisition scheme. (a) Sketch of the STED-ISM custom microscope. The FPGA board controls two excitation lasers, a depletion laser, a couple of galvanometric mirrors, and a piezo-electric-controlled stage. The same FPGA board reads the output of each channel of the SPAD array detector. (b) Working principle of the heterodyne acquisition. The lifetime decay is sampled along multiple excitation periods using a detuned sampling frequency. The sampling accumulates a delay in each period, enabling data collection at different time points. Each sampling period is subsampled by short windows, boosting the temporal resolution.
The DFD-TDC performs digital heterodyning: excitation and sampling frequencies, and , are slightly different. The periodicity of the beating signal determines the duration of a DFD cycle. This latter spans multiple excitation cycles, time-tagging photons at different delays from the excitation pulse. In more detail, each sampling period is divided into an integer number of short windows at frequency . Thanks to a pair of counters (the phase and window indices), the photon arrival times are remapped into a single excitation period, generating a densely sampled histogram of the fluorescence decay with a temporal resolution beyond that of conventional sampling [Fig. 1(b)].
We designed our system to generate the excitation and sampling frequencies from the same FPGA clock: the excitation signal triggers the laser pulses, and the temporal histogram is fully calculated at the FPGA level. In short, our system embeds the control of the microscope and the measurement of the photon arrival time histogram in a single compact device. Advantageously, our architecture uses only a limited amount of hardware and software resources (see Note 1 in the Supplementary Material).
In this work, we use two specific configurations, running at and . Both implementations achieve a temporal resolution (i.e., the temporal width of the histogram bin) of about 400 ps. Instead, the temporal precision is dictated by the value of , which is about 2 ns.
2.1 Fluorescence Lifetime Assay
To validate the proposed architecture, we used the DFD configuration running at to measure the photon arrival time histogram [Fig. 2(a) top] from a sample of diluted solution of Alexa 594 with a well-known fluorescence lifetime ().28 Given the photon arrival time histogram, we can estimate the fluorescence lifetime value by fitting the decay to an exponential function convolved with the system impulse response function (IRF). Nonetheless, the phasor approach enables a fit-free lifetime value estimation, inherently considers multiexponential decays, and grants access to a straightforward graphical representation of lifetime values.29 Thus we use the photon arrival time histogram to calculate the intensity normalized phasor vector components for the first harmonic and the corresponding phase and modulation values . We then represent the phasor vector for the Alexa 594 into a two-dimensional histogram called the phasor plot [Fig. 2(a) top, inset]. The theoretical model (see Note 2 in the Supplementary Material) predicts that the phasor of single-exponential decay lies on a semicircle in the complex plane, known as the universal circle.30 However, the IRF of the system alters the experimentally measured decay, whose phasor no longer lies on the universal circle. Starting from the reference lifetime value of the Alexa 594, we calculated the corrected phasor. The ratio of the measured to the corrected phasor leads to the phasor components of the system’s IRF, which can be used to calibrate all successive measurements (see Note 2 in the Supplementary Material). Indeed, we measured a solution of Rhodamine 101, whose raw and corrected phasors are shown [Fig. 2(a)]. The fluorescence lifetime calculated from the phase of the calibrated phasor phase value is , similar to the values reported in the literature.31 In our DFD implementation, the phase difference between the excitation and the sampling signal is not fixed and may vary from measurement to measurement. Thus we need to measure the phase of an additional reference. For this, we record the trigger signal used to synchronize the excitation laser as an additional virtual channel. The calibration procedure is applied independently to each element of the SPAD array detector (Fig. S2 in the Supplementary Material).
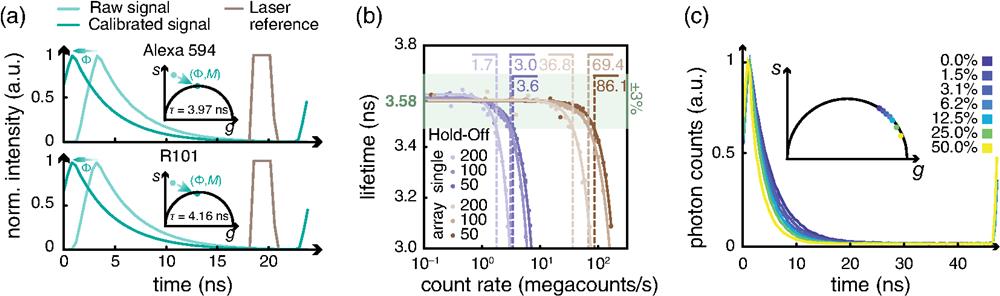
Fig. 2. Validation of the DFD architecture. (a) Calibration procedure. The measured phasor is shifted and rescaled by the IRF calibration values. Additionally, each histogram is phase-shifted by the value provided by the trigger reference signal. Top, calibration of Alexa 594; bottom, calibration of Rhodamine 101. (b) Measured lifetime of an autofluorescent plastic slide as a function of the detected photon flux and of the hold-off time of the SPAD. The reported values are the saturation thresholds (in units of megacounts/s), defined as the flux at which lifetime estimation deteriorates by a factor larger than 3%. (c) Lifetime of a fluorescein solution at increasing concentration of a quencher, potassium iodide. We depict with the same colors the exponential curves and the corresponding phasors.
Implementing the TDC using a DFD approach allows for recording virtually all photon signals. Indeed, the dead time of the DFD module is given by the window duration, in our implementation . Within each window, the DAQ system can only discriminate between the presence or absence of the detection event. Nonetheless, state-of-the-art SPAD array detectors have hold-off times in the range of tens of nanoseconds. Thus the dead time of the DFD module does not introduce any additional practical limitation. However, the low speed of the USB 2.0 interface, which connects the card to the computer, constrains the data-transfer rate of our implementation of the multichannel DFD-DAQ system. Assuming a depth of 16 bits for each time bin of the histogram and 26 parallel channels, USB 2.0 is able to transfer the whole histogram every , imposing a lower limit on the pixel dwell time.
We experimentally verified the maximum photon flux that can be sustained by our multichannel DFD-DAQ module. We registered the fluorescence signal generated by an autofluorescent plastic slide (Chroma) at increasing excitation power. We calculated the fluorescence lifetime from the histograms retrieved at increasing photon fluxes [Fig. 2(b)]. The fluorescence lifetime of the slide does not depend on the excitation power. Thus a reduction of the estimated fluorescence lifetime value indicates the saturation of the DAQ system. Indeed, at high count rates, the early photons are most likely to be detected because of the hold-off of the detector. This phenomenon, known as the pile-up effect, causes the appearance of an additional short-lived exponential component and a decrease of the measured lifetime. In these experiments, the photons are not distributed uniformly across the whole detector array, but according to the diffraction laws. Specifically, light spreads according to the fingerprint distribution,10 namely, a bell-shaped function peaked in the central element of the detector array. The central element starts saturating (namely, the lifetime error is ) above . To measure the saturation of the whole system, we integrated all photon arrival histograms before calculating the fluorescence lifetime. We find the saturation threshold of the whole system to be at 36.8 megacounts/s. The dead time of the SPAD elements is the major cause of saturation. Thus reducing the hold-off time of the SPAD array from 200 to 50 ns, the saturation threshold shifts toward higher photon fluxes: for the central element and 86.1 megacounts/s for the whole system.
Finally, we used our platform to measure the fluorescence lifetime values for a series of fluorescein solutions obtained with increasing concentrations of the quencher potassium iodide. As expected, the platform retrieves shorter lifetimes as the quencher concentration increases [Fig. 2(c)].
2.2 FLISM for Functional Imaging
After validating the multichannel DFD-DAQ module, we used the module to implement FLISM.6,9 FLISM harnesses the photon-resolved spatiotemporal information provided by the SPAD array to obtain a superresolved fluorescence lifetime map of the sample. On each acquisition, the microscope builds a 5D photon-counts map . We reconstruct a superresolution image from the raw data using adaptive pixel reassignment (APR), as in conventional ISM. In FLISM, we iterate the APR analysis on each temporal bin, obtaining a time-resolved superresolution image . We apply phasor analysis to the latter to obtain the fluorescence lifetime map . We started by imaging fixed HeLa cells, in which we stained Tubulin with ATTO 647N [Fig. 3(a)] and nuclear-pore complexes with Abberior STAR 580 [Fig. 3(b)]. We used the excitation wavelength and detection filter that best matches with the probes. For both structures, FLISM outperforms conventional confocal laser scanning microscopy. The FLISM images show a better resolution than the open-pinhole confocal microscopy counterpart (Figs. S3 and S4 in the Supplementary Material). When closing the pinhole, the optical resolution of confocal microscopy should reach the FLISM resolution.6 However, the pinhole rejects most of the fluorescence photons, drastically reducing the SNR of the image. The SNR is a crucial parameter for fluorescence lifetime imaging. Regardless of the methodology employed to analyze histogram arrival times, a lower SNR directly corresponds to a less precise lifetime measure. The differences in fluorescence lifetime precision among the three imaging modalities (open-pinhole, closed-pinhole, and ISM) are evident when comparing the respective fluorescence lifetime maps and the phasor plots [Figs. 3(a) and 3(b) and Fig. S5 in the Supplementary Material]. Since the tubulin network and the nuclear-pore complexes are stained in fixed cells, the fluorescence lifetime should be constant across the whole sample. A potential source of lifetime heterogeneity in the sample could be the high concentration of fluorophores: in this condition, fluorophores can self-quench each other, resulting in shorter lifetime values.27 Here we assume that self-quenching and similar phenomena are negligible. Thus we ascribe the variance of fluorescence lifetime values to estimation errors. Closed-pinhole images show different pixels whose phasors do not lie on the universal circle. Indeed, the poor SNR of confocal images hinders a reliable lifetime estimation, resulting in low precision and low accuracy. In contrast, FLISM images provide better results, as demonstrated by the narrower phasor cloud well centered on the universal circle. Comparing the fluorescence lifetime histograms confirms the same trend. Closed-pinhole confocal microscopy shows a broader distribution than open-pinhole confocal microscopy and ISM (Fig. S5 in the Supplementary Material). The effect, known as super-brightness,32 makes the SNR of the ISM image even better than that of open-pinhole confocal microscopy, further enhancing the precision of FLISM.
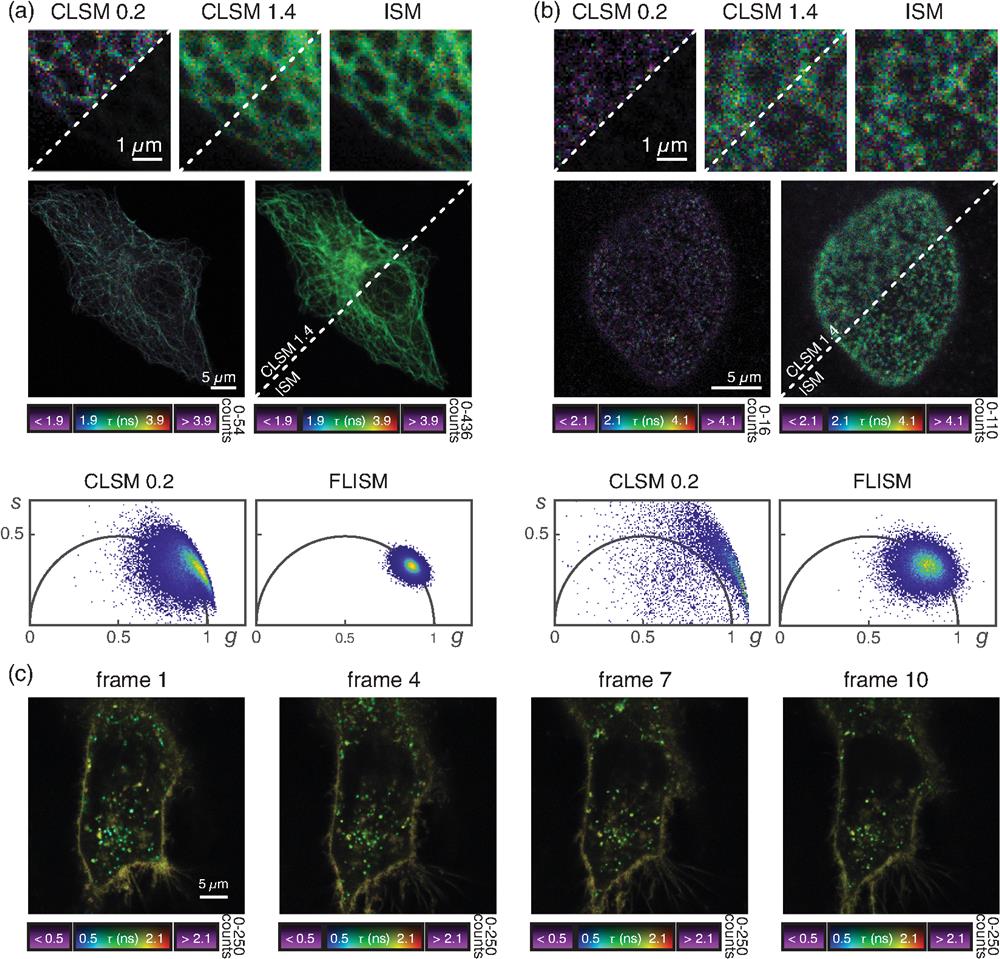
Fig. 3. FLISM imaging. (a) Alpha-tubulin stained with ATTO 647N in fixed HeLa cells. (b) Nuclear pore complexes stained with Abberior STAR 580 in fixed HeLa cells. (c) Live HeLa cell labeled with the CellBrite® NIR 680 cytoplasmic membrane dye. The displayed frames are captured at
Next, we used the FLISM approach to measure living HeLa cells labeled with the CellBrite® NIR 680 cytoplasmic membrane dye. Because the dye uniformly labels any membrane structure, conventional intensity-based imaging cannot distinguish the signal stemming from cell membrane or membrane-enclosed organelles (e.g., lysosomes). Conversely, FLISM reveals a broad distribution of the fluorescence lifetime values, which allows for distinguishing the plasma membranes (longer fluorescence lifetime) from intracellular vesicles or lipid-based structures (shorter fluorescence lifetime). Indeed, the variation of the probe fluorescence lifetime allows for monitoring the changes in the local environment. The superior SNR of ISM imaging allows for working at reduced illumination intensity, thus allowing for long-term imaging [Fig. 2(c)]. We follow the variations of the fluorescence lifetime of the membrane dye within the HeLa cells for more than 2 h.
2.3 FLISM for Multispecies Imaging
Biological investigations often require visualizing multiple biomolecules and cellular compartments simultaneously. Multispecies imaging generally targets the different biomolecules with spectrally separable fluorophores. A time-resolved detection system enables separating fluorophores based on their fluorescence lifetime, offering a viable alternative to distinguishing dyes with similar emission and excitation spectra. Conveniently, this solution requires only a single detector. Furthermore, lifetime separation does not preclude spectral separation. Indeed, the two approaches can be combined to enable the separation of an even wider number of species.20
Here we demonstrate how our DFD-DAQ module can implement imaging through fluorescence lifetime multiplexing. In particular, we show that FLISM method can perform superresolution multispecies imaging using a single SPAD array detector. We stained -tubulin and lamin A in fixed MCF10A cells with Abberior STAR RED and Abberior STAR 635, respectively. The two dyes show almost identical excitation spectra and largely overlapping emission spectra. We excited both dyes with the same pulsed laser () and detected their fluorescence signals within the same spectral window and SPAD array detector. However, the two probes emit fluorescence with substantially different lifetimes. A straightforward approach for fluorescence lifetime multiplexing imaging is phasor segmentation. Typically, two (or more) regions on the phasor plot are identified using the single-probe samples. The same regions are then reported in the phasor plot of the multiprobe image, and the points on these regions are backprojected to generate the single-species images. The phasor segmentation method is easy to use, fast, and visually intuitive. However, it does not fully extract the information embedded in the phasor plot. Indeed, phasor segmentation assigns each pixel to a single fluorophore—regardless of how many components are present in the probed region—leading to poor results in many practical scenarios.
For this reason, we analyzed FLISM data with a linear unmixing algorithm—called phasor separation—inspired by previously reported unmixing methods.25,33,34 This latter decomposes the FLISM signal of each scan point into the components associated with the two species. This method exploits the phasor representation of fluorescence decays. We describe each decay with the first frequency component of the corresponding Fourier transform—namely, the phasor. With such an approach, the mixed signal is conveniently represented as the linear combination of two phasors, one for each fluorophore. Thus we transform a nonlinear regression algorithm—such as a model-based fitting—into a linear problem that admits an easy-to-compute solution (see Note 3 in the Supplementary Material). We tested the phasor separation method on the FLISM data set of MCF10 cells. First, we apply the phasor analysis to the images of two separate samples containing only one or the other dye. Thus we retrieve the corresponding phasor coordinates, which map to different points of the universal circle. Then we compared the phasor segmentation and the phasor separation method. Both methods are feasible, but phasor segmentation results in an excessively sharp classification that does not admit the presence of multiple components in a single pixel. Instead, the phasor separation method provides a smoother fluorophores separation, reweighting each pixel by the corresponding fractional components [Fig. 4(a) and Fig. S6(a) in the Supplementary Material].
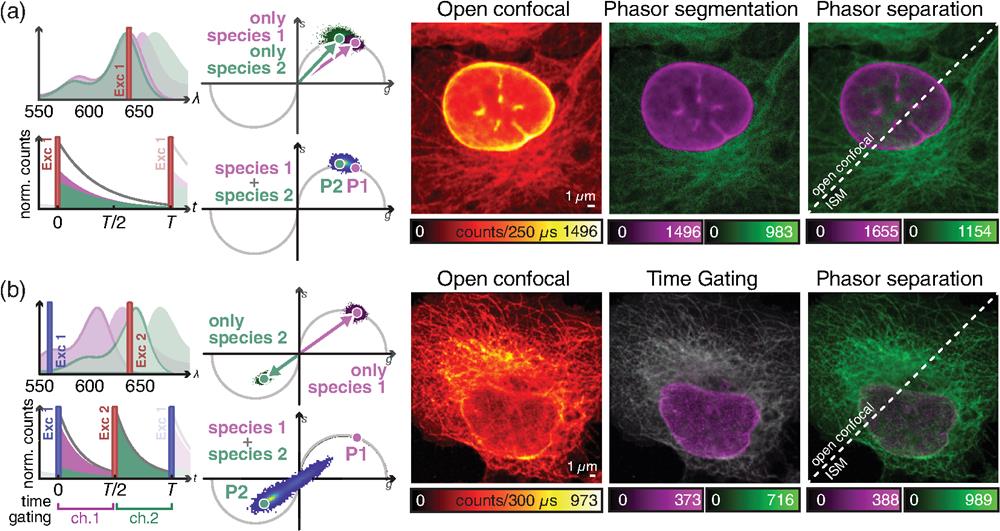
Fig. 4. Time-resolved acquisition for multicolor ISM imaging. (a) ISM imaging of different species, reported by two dyes with similar “spectra” but different fluorescence lifetimes. (b) Pulse-interleaving ISM imaging of different species with overlapping absorption spectrum and identical lifetime. Left, schematic diagrams of (top) the absorption/emission spectra of fluorophores 1 (magenta) and 2 (green), and (bottom) of the detected signal over time. The excitation lines are also represented. Right, phasor representation of (top) two measurements obtained with only one species labeled at a time, and (bottom) of the measurement with both species labeled. Left image, raw open confocal image. Central image, open confocal result obtained by (a) segmenting the phasor space in two regions, related to the first and the second fluorophores or (b) by temporal gating of the detected signal in two temporal channels, related to the first and the second excitation events. Right image, open confocal and ISM results obtained by phasor separation.
An alternative approach for simultaneous multispecies imaging with a single SPAD array detector is the pulsed interleaved excitation (PIE) technique. The PIE technique can be applied if the probes have different absorption spectra but similar emission spectra. It consists of a sequential excitation at different wavelengths, synchronized with the signal recording. This strategy reduces the spectral emission cross talk without significantly reducing the acquisition speed, since the excitation repetition rate can be as fast as tens of megahertz. The PIE technique encodes the spectral information into the time domain, which can be used to separate the different fluorophores if no other discrimination is possible. Indeed, the PIE technique is key when the dyes have very similar lifetime values, so they cannot be reliably separated using this latter. In more detail, a phasor associated with the second excitation window will appear rotated by 180 deg in the complex plane. This approach well conditions the phasor unmixing problem well, which would be ill-posed if the lifetime value of the dyes is the same. We achieve PIE by synchronizing several pulsed lasers with different colors. The pulses from the different lasers are alternated with a delay much longer than the fluorescence lifetime of the probe to ensure that a dye emits all the photons before exciting the other dye. We record the photons’ arrival time information with respect to the excitation pulse, enabling the generation of different images obtained by binning the photons in different temporal windows, one for each species.35 This method, known as time-gating, fails if the absorption spectra of the probes overlap, since one fluorophore could be excited by multiple excitation colors. Harnessing the phasor information, we remove the cross talk by unmixing the contributions of different fluorophores by decoding the absorption information encoded in the fluorescence dynamics. To demonstrate the feasibility of the proposed approach, we performed PIE imaging of a fixed HeLa cell (Tubulin labeled with ATTO 647N and nuclear-pore complexes with Abberior STAR 580). The two dyes have an overlapping absorption spectrum. Thus time-gating alone cannot effectively separate the structures labeled with the two dyes. In particular, ATTO 647 can be excited by the lasers at 560 and 640 nm, contributing to the counts of the window assigned to STAR 580. Additionally, the two dyes have very similar lifetime values (nominally, both have ). Thus the two channels cannot be separated using the lifetime information from a single excitation cycle. To calculate the phasor components of each species, we first imaged two samples with individual labeling. Then we calculated the phasors along the complete excitation period (). Thus the photon-arrival histogram contains the fluorescence excited by the green and red pulses. The phasor of STAR 580 (excited by the laser at 560 nm) is localized in the universal semicircle of the first quadrant of the complex plane. Indeed, STAR 580 cannot be excited by red light and emits fluorescence with single exponential decay. The dye ATTO 647N is primarily excited by the red laser but also partially excited by the green laser. The corresponding fluorescence decay is the linear composition of two exponential decays separated in time by half of the excitation period. Since the second excitation, caused by the green laser, is more effective than the first one, the phasor is localized in the third quadrant of the complex plane. Thus the absorption cross talk is naturally encoded in the phasor plots of the two species. The phasor separation method can exploit such information to separate the two species, even in the presence of strong cross talk. For comparison, we also show the results of simple time-gating, in which one of the two channels of the image contains mixed contributions. As expected, time-gating cannot preserve the specificity of the labeling if the dyes have overlapping absorption spectra. We also show the unmixing performed on the individual time windows. Since the two dyes decay with the same lifetime, they cannot be distinguished from a single excitation cycle, and the separation fails (Fig. S7 in the Supplementary Material). In contrast, the phasor separation method successfully distinguishes the contributions of each dye [Fig. 4(b) and Fig. S6(b) in the Supplementary Material].
2.4 FLISM for Nanoscopy Imaging
Another advanced fluorescence LSM technique that can significantly benefit from the single-photon imaging paradigm is STED microscopy. In STED microscopy, a second laser beam, the STED beam, induces stimulated emission on the fluorescent probe. The STED beam is engineered in phase and polarization to generate a doughnut-shaped intensity distribution at the focus. By spatially overlapping the foci of the depletion and excitation beam, the probed region of the laser-scanning microscope reduces in size well below the diffraction limit: the higher the STED beam intensity is, the smaller the probed region is.36 The combination of STED microscopy with ISM enables a reduction of the STED beam intensity to achieve a target resolution. This benefit is maximal at low STED beam intensity and vanishes for high-intensity values. However, live-cell imaging is typically performed at reduced STED beam intensity to mitigate phototoxicity. Indeed, we recently demonstrated that live-cell superresolution imaging is feasible for extended time-lapse experiments without damaging the sample. Furthermore, the additional spatial information that the detector array provides enables effective background removal.10
In the context of gentle live-cell STED microscopy, a widely used approach to reduce the STED beam intensity is time-resolved STED microscopy27,37,38—now implemented in all commercial instruments. Time-resolved STED microscopy is a class of implementation that leverages the relation between fluorescence depletion and fluorescence lifetime. Indeed, stimulated emission opens a new relaxation pathway for the probe, whose rate is proportional to the intensity of the STED beam. Thereby, the higher the intensity is, the higher the efficiency of depletion is, and the shorter the fluorescence lifetime of the probe is. Because the STED beam intensity at the focus is shaped like a doughnut, it induces a fluorescence lifetime spatial signature: the fluorescence lifetime is the shortest at the periphery and unperturbed at the center of the probed region, where the intensity of the depletion beam is at its maximum and minimum, respectively. Time-resolved STED microscopy harnesses the spatial dependency of fluorescence lifetime to distinguish the fluorescence signal generated from the center or periphery of the probed region. The result is a smaller effective point spread function (PSF). Thus the resolution is enhanced without increasing the STED beam intensity.
Time-resolved STED microscopy initially used a time-gated detection37,38 to remove the short-lived fluorescent signal. The fluorescence signal is recorded only after a fixed delay (a fraction of the probe’s natural fluorescence lifetime) from the excitation events triggered by the pulsed laser. The longer the delay is, the smaller the effective probed region is. However, time-gated detection also rejects part of the fluorescence signal generated from the center, thus reducing the SNR. Given the photon arrival time histogram of the probed region, a computational alternative can solve the hardware time-gated detection limitations, providing the same resolution improvement but without a strong SNR reduction. In particular, the SPLIT method can separate the long-lived and short-lived fluorescence signal generated from the inner and outer parts of the STED microscopy probed region, respectively.25,26,39,40 The SPLIT method represents the two decays as a linear combination of phasors. Thus the unmixing problem of the different components is solved by the SPLIT method by inverting a simple linear system (see Note 3 and Figs. S8 and S9 in the Supplementary Material).
Here we used our proposed DFD-DAQ module to introduce time-resolved STED-ISM. In particular, we combined APR-based STED-ISM with the SPLIT approach. We call this combination SPLIT-STED-ISM. We first applied the SPLIT approach on the 5D raw STED photon-counts map , to obtain a set of images with increased resolution. Then we used the APR method to achieve the final SPLIT-STED-ISM result, with further increased resolution and SNR with respect to the original raw STED measurement. The APR algorithm calculates the shift vectors directly from the data, thus adapting toward the power of the STED beam. Notably, the shift vectors retrieved before and after applying the SPLIT algorithm differ due to the effective separation of the early and late photons (Fig. S10 in the Supplementary Material). Thus resolution and SNR are maximized if APR is applied after SPLIT.
We tested the new SPLIT-STED-ISM method on fixed HeLa cells with ATTO 647N-labeled tubulin. The longer the duration of the STED pulse is, the more useful SPLIT is in removing the incomplete depletion background at the periphery of the probed region.27 In a practical scenario like ours, the pulse width is nonnegligible with respect to a probe’s fluorescent lifetime—about 600 ps to a few nanoseconds. Thus combining STED-ISM with SPLIT produces a high-contrast and high-quality image [Fig. 5(a) and Fig. S11(a) in the Supplementary Material]. Finally, we complemented nanoscopy with multispecies imaging. In detail, we used the PIE approach combined with STED microscopy. We separated the different fluorophores (STAR RED for nuclear pore complexes and STAR ORANGE for the Golgi apparatus) through time-gating—given the lack of fluorophores cross talk. Thus we demonstrated how, by encoding the spectral information into the time domain, we enable multicolor STED imaging using a single detector. Multispecies imaging does not hinder the SPLIT method, which can be applied to each excitation window. Thus we generated a multicolor SPLIT-STED-ISM image [Fig. 5(b) and Fig. S11(b) in the Supplementary Material]. With a single measurement, we separated the contribution of different fluorophores, performed superresolution imaging of the labeled structures, and improved contrast and resolution of each channel (Figs. S12 and S13 in the Supplementary Material). This result demonstrates the vast capabilities of our photon-resolved platform. Indeed, the new possibilities unlocked by the additional temporal and spatial information do not compromise any of the conventional features of STED imaging. We achieved each benefit with a simple tailored acquisition system based on a SPAD array detector without changing the core optical architecture or the experimental protocols.
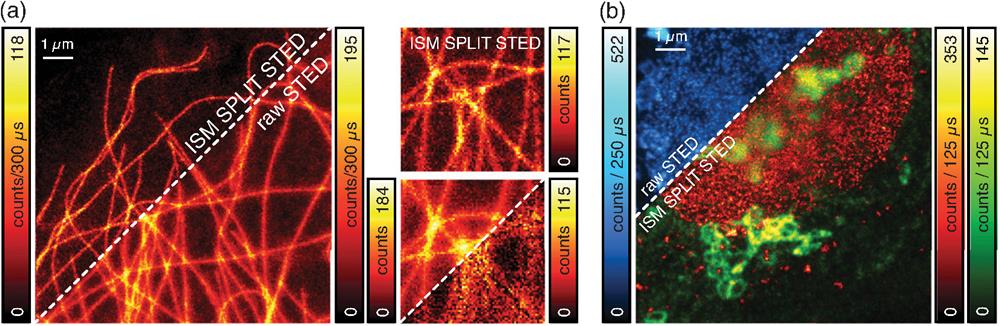
Fig. 5. SPLIT-STED-ISM. (a) Side-by-side comparison of raw STED imaging and SPLIT-STED-ISM imaging of the cytoskeleton network. The insets show magnified details of the network. The incomplete depletion background is also visualized (bottom inset, right corner). (b) Multispecies SPLIT-STED-ISM imaging (green, Golgi apparatus; red, nuclear pore complex protein, NPC). Pulse-interleaving excitation and time-resolved detection enable the separation of two color channels—one per each fluorophore—and to apply SPLIT to the NPC channel. The upper corner shows the raw STED image (blue), while the lower corner shows the final multi-color SPLIT-STED-ISM image.
3 Discussion
We presented an FPGA-based multichannel DAQ system tailored for fluorescence LSM with asynchronous read-out SPAD array detectors. This system uses the DFD principle to implement 25 low-resource TDCs. Using limited resources of the FPGA, our architecture enables the integration of the DAQ-DFD module and the microscope control unit into the same FPGA-based board, greatly reducing the microscope cost and complexity. These benefits come at the cost of a lower temporal precision and sampling ( and , respectively) than other FPGA-based TDC architectures, which recently have been shown to reach 10 ps precision.41 Nonetheless, the specifications of our architecture are fit to implement FLISM, making its numerous applications easily accessible. On the other side, a limitation of the current DAQ module implementation is the poor maximum data transfer rate of the USB 2.0 protocol, which constrains the pixel dwell time of the scanning microscope to . This problem can be solved using a board with similar FPGA but faster data transfer protocols (e.g., USB 3.0 or PCI Express). This board would also open to more channels (e.g., 49 channels), since the bottleneck is not the FPGA resource but the limited data transfer rate.
We used the proposed DFD-DAQ system to implement the well-established FLISM technique and demonstrate novel advanced imaging techniques based on the single-photon laser scanning microscopy paradigm. In particular, we demonstrated a method capable of distinguishing fluorophores on the base of their absorption spectra. We used the pulse-interleaving excitation scheme to encode the spectral information of the fluorophores into the time domain. Successively, we used a phasor-based approach to decode the fluorescence light emitted by the two species. The proposed method automatically corrects the fluorophore excitation cross talk. Notably, the same approach can also distinguish fluorophores with different lifetimes, setting the basis for multispecies ISM imaging with a single detector. Thus we effectively increased the portfolio of usable dyes.
We also demonstrated the combination of ISM with time-resolved STED microscopy. In a recent paper, we showed the benefits of combining STED microscopy with the APR method to reduce the STED beam intensity, thus minimizing the risk of inducing photodamage.10 Here we demonstrate that leveraging single-photon temporal information can further boost the benefits. We proved that the SPLIT method increases the resolution and contrast of the image. Since the algorithm can be applied to each pixel and channel independently, it leaves the dimensionality of the ISM data set intact. Thus the processed data are still compatible with the recent and advanced image reconstruction algorithm developed for ISM, such as focus-ISM10 or multi-image deconvolution.42 We envision that maximum likelihood reconstruction methods that consider both the spatial and temporal information will emerge as a superior reconstruction tool for time-resolved STED-ISM data sets. Indeed, the spatiotemporal information is encoded in the temporal PSFs: the stimulated emission process introduces a temporal evolution on the effective PSF. Namely, the stronger the effect of the STED beam, the narrower the PSF associated with each scanned image (Figs. S10 and S14 in the Supplementary Material).
In conclusion, we believe that the proposed architecture will make photon-resolved image scanning microscopy easily accessible, paving the way for gentle, versatile imaging at high spatial resolution and information content.
4 Methods
4.1 Microscope Architecture
For this work, we updated the STED-ISM setup described previously,10 adding the possibility of exciting the sample also with a green laser beam and recording the photon arrival time histograms (Fig. S1 in the Supplementary Material). Briefly, the excitation beams were provided by two triggerable pulsed ( pulse width) diode lasers (LDH-D-C-640 and LDH-D-C-560, Picoquant) emitting at 640 and 560 nm, respectively. The STED beam was provided by a triggerable subnanosecond ( pulse width) pulsed fiber laser (Katana 775, OneFive) emitting at 775 nm. We adjusted the laser power for all beams using their respective drivers and control software. For the 640 and 516 nm laser beams, we can further modulate and adjust the power using acoustic optical modulators (AOMs, MT80-A1-VIS, AA Opto Electronic), which also act as fast shutters. All laser beams are coupled into a different polarization-maintaining fiber (PMF) to transport the beams to the microscope easily. In all cases, we used a half-wave plate (HWP) to adjust the beam polarization parallel to the fast axis of the PMF. For the STED beam, we used a PMF designed explicitly for high-power applications (QPMJ-A3HPM, 3S-633-4/125-3-7-1, AMS Technology). The STED beam emerging from the PMF is collimated, filtered in polarization by a rotating Glan–Thompson polarizing prism and phase-engineered through a polymeric mask imprinting helical phase-ramps (VPP-1a, RPC Photonics). We rotated a quarter-wave plate and an HWP to obtain circular polarization of the STED beam at the back aperture of the objective lens. A set of dichroic mirrors allows the combination of all laser beams (i.e., excitation and STED) and the separation of the fluorescence signals. Two galvanometer scanning mirrors (6215HM40B, CT Cambridge Technology), a scan lens, and a tube lens—of a commercial confocal microscope (C2, Nikon)—deflect and direct all the beam toward the objective lens (CFI Plan Apo VC , 1.4 NA, Oil, Nikon) to perform the raster scan on the specimen. The fluorescence light was collected by the same objective lens, descanned, and sent to the detector. A 300 mm aspheric lens (Thorlabs) focuses the fluorescence light into a completely open pinhole plane generating a conjugated image plane with a magnification of . A telescope system, built using two aspheric lenses of 100 and 150 mm focal length (Thorlabs), conjugates the SPAD array with the pinhole and provides an extra magnification factor. The final magnification on the SPAD array plane is ; thus the size of the SPAD array projected on the specimen is (at the far-red emission wavelength, i.e., 650 nm). Two notch filters (561 and 640 nm) and a short-pass filter (720 nm) reject the light from the laser beams reaching the image plane (i.e., before reaching the SPAD array detector). Depending on the experiments, fluorescence light can be selected using a bandpass filter (590/50 nm and/or 685/70 nm). The setup mounts a asynchronous read-out SPAD array detector (PRISM-light kit, TTL version, Genoa Instruments). Every photon detected by any of the 25 elements of the SPAD array generates a TTL signal that is delivered through the dedicated channel (one channel for each sensitive element of the detector) to a multifunction FPGA-based I/O device (NI USB-7856R from National Instruments). The board acts both as DFD-DAQ system and control unit. The LabVIEW-based software, inspired by the Carma software,43 implements the DFD and microscope control modules. Specifically, the software controls all the microscope devices needed during the image acquisition, such as the galvanometric mirrors, the axial piezostage, and the AOMs, builds the 5D photon-counts map , and visualizes the images (e.g., the image obtained from the integration along the and coordinates). Furthermore, the control unit provides the three synchronized signals for triggering the excitation and the STED beams (i.e., ). Each synchronization signal is sent to a different picosecond delayer to implement the pulse sequence for pulse-interleaving excitation and for an efficient stimulated depletion process. When necessary, the delayer also converts the TTL from the control unit to a NIM signal for the laser driver.
4.2 Experimental Procedure
To respect the Nyquist–Shannon criterion, the sampling frequency has to be larger than twice the largest frequency of the signal. For an exponential decay, the cut-off frequency is [see Eq. (26) in the Supplementary Material]. Thus the sampling window should be smaller than . Such a criterion is always respected for the parameters of our DFD implementation () and a typical fluorophore decay (a few nanoseconds). Furthermore, the DFD heterodyne measurement enables a reduction of the bin size down to (see Note 1 in the Supplementary Material). To collect the full fluorescence decay, the excitation period has to be larger than . In the case of pulse-interleaving excitation, the same condition applies to each time window. In general, , where is the number of excitation windows. In the current implementation, or and or , ensuring the exponential decay is not cropped in the acquisition process. Thus we ensure that no data distortion happens in any of the performed measurements.
4.3 Sample Preparation
4.3.1 Quenched fluorescent solutions
For spectroscopy measurements [Fig. 2(c)], we prepared different solutions of fluorescein (46955, free acid, Sigma-Aldrich, Steinheim) at different concentrations of the quencher salt potassium iodide [60399-100G-F, BioUltra, (AT), Sigma-Aldrich]. We first dissolved fluorescein from powder into DMSO (Sigma-Aldrich), and we further diluted it to a 1:1000 (mass-to-volume ratio) by adding ultrapure water. Then we diluted the solution at different volume ratios with the potassium iodide quencher (1:2, 1:4, 1:8, 1:16, 1:32, and 1:64). All samples were made at room temperature. A new sample solution was prepared before each measurement—nevertheless, not optimal; we excited the sample using the 560 nm laser beam.
4.3.2 Fluorescent beads
To qualitatively characterize the spatial resolution enhancement of ISM and STED microscopy (Figs. S3 and S8 in the Supplementary Material), we used a commercial sample of ATTO 647N fluorescent beads with a diameter of 23 nm (Gatta-BeadsR, GattaQuant).
4.3.3 Fixed cell imaging
To validate our DFD-DAQ system for FLISM imaging [Figs. 3(a) and 3(b), Fig. S4 in the Supplementary Material], multispecies imaging based on pulse-interleaving excitation [Fig. 4(b)], and single-color STED imaging [Fig. 5(a)], we used a fixed HeLa cell labeled to visualize -tubulin and Nup 153 (nuclear pore complexes). HeLa cells were seeded on coverslips in a 12-well plate (Corning Inc., Corning, New York, United States) and cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco™, ThermoFisher Scientific) supplemented with 10% fetal bovine serum (Sigma-Aldrich, Steinheim, Germany) and 1% penicillin/streptomycin (Sigma-Aldrich) at 37°C in 5% . After 24 h, cells were incubated in a solution of 0.3% Triton X-100 (Sigma-Aldrich) and 0.1% glutaraldehyde (Sigma-Aldrich) in BRB80 buffer (80 mM Pipes, 1 mM EGTA, 4 mM , pH 6.8, Sigma-Aldrich; 1 M = 1 mol/L) for 1 min. After fixation with a solution of 4% paraformaldehyde (Sigma-Aldrich) and 4% sucrose (Sigma-Aldrich) in the BRB80 buffer for 10 min, cells were washed 3 times for 15 min in phosphate-buffered saline (PBS, Gibco™, ThermoFisher Scientific). Cells were incubated in a 0.25% Triton-X-100 solution in BRB80 buffer for 10 min, then washed 3 times for 15 min in PBS. After 1-h incubation in blocking buffer [3% bovine serum albumin (BSA, Sigma-Alrich) in BRB80 buffer], fixed HeLa cells were incubated with monoclonal mouse anti--tubulin (1:1000, Sigma-Aldrich) and rabbit anti-Nup153 (1:500, ab171074, Abcam, Cambridge, United Kingdom) antibodies diluted in the blocking buffer for 1 h at room temperature. Anti--tubulin and anti-Nup153 antibodies were, respectively, revealed by ATTO 647N goat anti-mouse IgG (1:400, Sigma-Aldrich) and STAR 580 goat anti-rabbit IgG (1:400, Abberior GmbH, Göttingen, Germany) diluted in BRB80 buffer. After 1 h of incubation, cells were rinsed 3 times in PBS for 15 min. Finally, coverslips were mounted onto microscope slides (Avantor, VWR International, Milano, Italy) with 2,2-thiodiethanol (Sigma-Aldrich).
To validate our DFD-DAQ system for multispecies imaging based on fluorescence lifetime [Fig. 4(a)], we used fixed MCF10A labeled to visualize -tubulin and lamin A. MCF10A cells plated on coverslips coated with the 0.5% (mass-to-volume ratio) pork gelatin (G2500, Sigma-Aldrich) were washed 3 times with prewarmed PBS, fixed with formaldehyde solution (3.7% methanol free) for 15 min, and washed 3 times with PBS. After the subsequent blocking with the blocking buffer composed of 0.1% Triton X-100 and 3% (mass-to-volume ratio) of BSA for 1 h, samples were incubated with the following primary antibodies for 1 h: mouse anti--tubulin (1:1000, T5168, Sigma-Aldrich) and rabbit anti-lamin A (1:1000, ab26300, Abcam). The samples were washed 2 times with the blocking buffer and incubated for 45 min in the dark with the following secondary antibodies: goat anti-mouse STAR RED (1:200, Abberior GmbH, Göttingen, Germany) and anti-rabbit STAR635 (1:200, Abberior GmbH, Göttingen, Germany). After washing the samples 5 times with PBS, they were mounted with the TDE mounting medium. Additionally, single staining experiments were performed in order to establish the fluorescence lifetime in the cell structure of interest (tubulin or lamin A) separately. All the steps were performed at room temperature, unless otherwise stated.
To validate our DF-DAQ system for multicolor STED-ISM imaging [Fig. 5(b)], we used a ready-to-image sample kit (Imaging set for STED at 775 nm, Abberior). The microscope slide contains fixed mammalian cells immuno-stained for a nuclear pore protein with STAR RED (Abberior GmbH, Göttingen, Germany) and the Golgi apparatus protein giantin with STAR ORANGE (Abberior GmbH, Göttingen, Germany).
4.3.4 Live cell imaging
For FLISM functional live-cell imaging, we used HeLa cells [Fig. 3(c)]. HeLa cells were seeded on -Slide 8 well (Ibidi GmbH, Gräfelfing, Germany) and cultured in DMEM (Gibco™, ThermoFisher Scientific) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin/streptomycin (Sigma-Aldrich) at 37°C in 5% . After 24 h, the growth medium was removed, and cells were incubated with CellBrite® NIR 680 (Biotium, Fremont, California, United States) for 20 min. After removing the staining medium, HeLa cells were rinsed 3 times with fresh growth medium; cells were incubated at 37°C for 5 min between each rinse. Finally, HeLa cells were imaged in live-cell imaging solution (ThermoFisher Scientific).
Giorgio Tortarolo got his MSc degree in engineering from the University of Genoa, Italy, in 2015. After graduating, he joined the Italian Institute of Technology, Genoa, Italy, as a PhD student, under the supervision of Dr. Giuseppe Vicidomini. He worked on an innovative image scanning microscopy platform and, in 2020 got his PhD. Currently, he is a Human Frontier Science Program cross-disciplinary fellow at EPFL, Lausanne, Switzerland, under the supervision of Prof. Suliana Manley.
Alessandro Zunino obtained his BSc degree in 2015 and his MSc degree in physics in 2018 from the University of Milan, Italy. In 2018, he started his PhD project at the Italian Institute of Technology (IIT), Genoa, Italy, under the supervision of Prof. Martí Duocastella and Prof. Alberto Diaspro. He obtained his PhD in physics in June 2022. Currently, he is a postdoctoral researcher within the group of Dr. Giuseppe Vicidomini at IIT.
Simonluca Piazza completed his MSc degree in biomedical engineering at the University of Genoa, Italy, 2014. He then earned his PhD in optical microscopy from the Italian Institute of Technology, Italy, 2018, under the guidance of Dr. Marti Duocastella, during which he developed advanced optical systems for imaging and fabrication. Since 2019, he has been a co-founder and CEO of Genoa Instruments, a company specializing in designing, manufacturing, and bringing to market super-resolution optical microscopes based on image scanning microscopy.
Mattia Donato got his BSc degree in 2010 and his MSc degree in 2013 in physics from the University of Genoa, Italy. His master thesis was developed in the Fusion Diagnostics Group at Uppsala University, Sweden, on the topic of neutron detectors characterization. Subsequently, he worked in the Detector Group at European XFEL focusing on the commissioning of an X-ray megahertz imager, obtaining his PhD from University of Hamburg in 2018. Currently, he works at Dr. Giuseppe Vicidomini's group, IIT, Genoa, Italy.
Sabrina Zappone studied medical biotechnologies at the University of Modena and Reggio Emilia, Italy. After graduation, she joined the Molecular Microscopy and Spectroscopy Group at the Istituto Italiano di Tecnologia, Italy, as a fellow student. Since November 2021, she has been a PhD student in bionanotechnology (bioengineering and robotics) at the University of Genoa, Italy, in collaboration with the Istituto Italiano di Tecnologia and under the supervision of Dr. Giuseppe Vicidomini.
Agnieszka Pierzyńska-Mach obtained her PhD in biophysics in 2017 from Jagiellonian University, Cracow, Poland. Subsequently, she completed a postdoctoral fellowship at the Italian Institute of Technology, Genoa, Italy, in the group of Prof. Alberto Diaspro and the European Institute of Oncology, Milan, Italy. In 2018, she secured an individual fellowship MSCA Horizon 2020 and investigated the nanoscale distribution of chromatin factors using advanced super-resolution microscopy techniques. Currently, she is an imaging scientist at the EPFL, Lausanne, Switzerland.
Marco Castello obtained his MSc degree in biomedical engineering from the University of Genoa, Italy, in 2012. Following his graduation, he earned his PhD in optical microscopy from the Italian Institute of Technology, IIT, Genoa, Italy, in 2017, under the mentorship of Dr. Giuseppe Vicidomini. He focused on advancing the field of optical microscopy with the development of a practical implementation of image scanning microscopy based on a SPAD array. Starting from 2019, he has served as co-founder and CTO of Genoa Instruments. The company focuses on the design and manufacturing of super-resolution optical microscopes based on image scanning microscopy.
Giuseppe Vicidomini obtained his PhD in computer science in 2008 from the University of Genoa, Italy, under the supervision of Prof. Alberto Diaspro. Following this, he joined the group of Prof. Stefan Hell at the Max Planck Institute in Goettingen, Germany. In 2011, he returned to Italy as a researcher at the Italian Institute of Technology, Genoa, and in 2016, he became the principal investigator of the Molecular Microscopy and Spectroscopy lab. Giuseppe Vicidomini is a grantee of the BrightEyes ERC project and co-founder of Genoa Instruments.
[14] C. J. R. Sheppard. Super-resolution in confocal imaging. Optik, 1988, 80: 53-54.
[15] C. B. Müller, J. Enderlein. Image scanning microscopy. Phys. Rev. Lett., 2010, 104: 198101.
Article Outline
Giorgio Tortarolo, Alessandro Zunino, Simonluca Piazza, Mattia Donato, Sabrina Zappone, Agnieszka Pierzyńska-Mach, Marco Castello, Giuseppe Vicidomini. Compact and effective photon-resolved image scanning microscope[J]. Advanced Photonics, 2024, 6(1): 016003.